Quantum vs Classical Computing in Chemistry: The Scaling Advantage for Drug Discovery and Materials Science
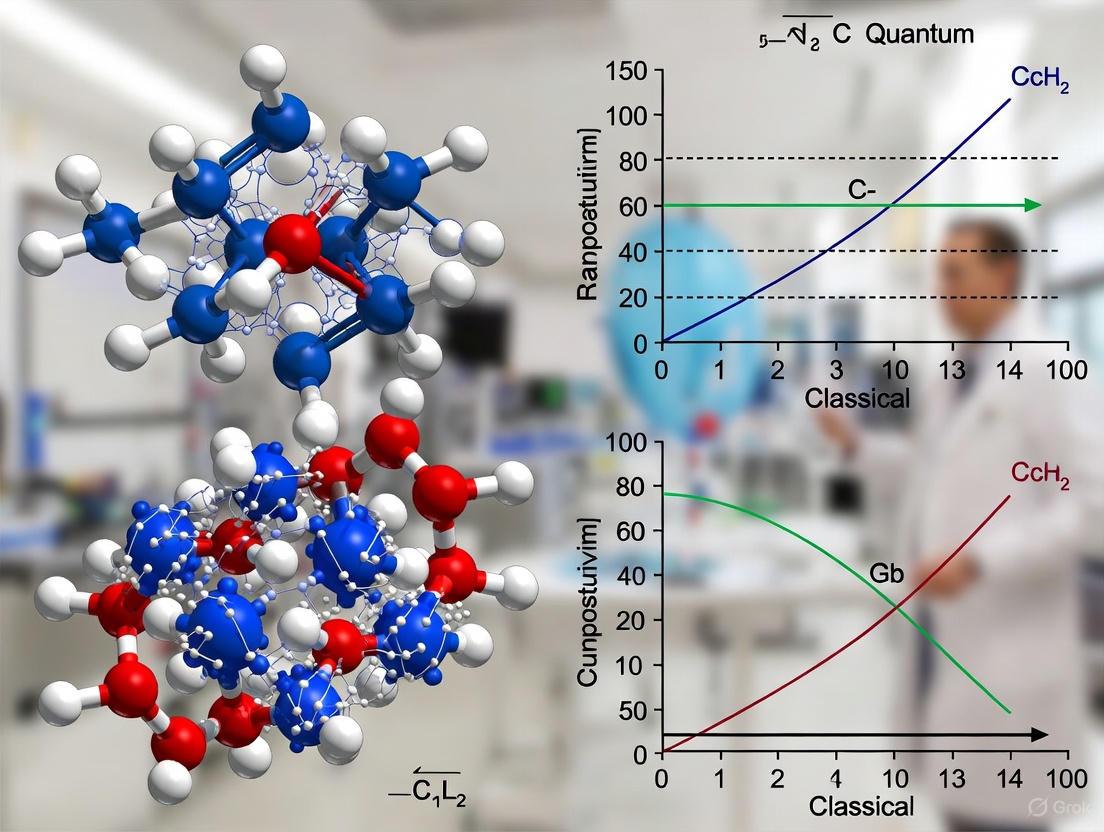
This article explores the fundamental computational scaling differences between quantum and classical computers in chemical simulations.
Quantum vs Classical Computing in Chemistry: The Scaling Advantage for Drug Discovery and Materials Science
Abstract
This article explores the fundamental computational scaling differences between quantum and classical computers in chemical simulations. Aimed at researchers and drug development professionals, it details how classical methods like Density Functional Theory (DFT) face exponential scaling limitations for complex quantum systems. In contrast, we examine how quantum algorithms, such as the Variational Quantum Eigensolver (VQE), offer a pathway to polynomial scaling, enabling the accurate simulation of molecular interactions, drug-protein binding, and catalytic processes that are currently intractable. The article provides a comparative analysis of current hybrid quantum-classical applications, discusses the critical challenges of error correction and qubit fidelity, and validates recent demonstrations of unconditional quantum speedup, ultimately outlining a future where quantum computing shifts chemistry from a field of discovery to one of design.
The Exponential Wall: Why Classical Computing Fails in Quantum Chemistry
In computational chemistry, the simulation of molecular systems is fundamentally limited by the scaling behavior of classical algorithms. The core of the problem lies in the exponential growth of computational resources required to solve the Schrödinger equation for quantum systems as their size increases. While classical computational methods such as Density Functional Theory (DFT) and coupled cluster theory have provided valuable approximations for decades, they inevitably face intractable complexity when modeling complex quantum phenomena like strongly correlated electrons, transition metal catalysts, and excited states [1].
Quantum computing emerges as a transformative solution to this scaling problem. Since molecules are inherently quantum systems, quantum computers offer a natural platform for their simulation, theoretically capable of modeling quantum interactions without the approximations that plague classical methods [1]. This comparison guide examines how quantum computational approaches are beginning to overcome the exponential scaling barriers that constrain classical methods in chemistry research, with particular relevance to drug development and materials science.
Computational Scaling: A Quantitative Comparison
The table below summarizes the fundamental scaling differences between classical and quantum computational methods for key chemistry simulation tasks.
Table 1: Scaling Comparison of Classical vs. Quantum Computational Methods
| Computational Method | Representative Chemistry Problems | Computational Scaling | Key Limitations |
|---|---|---|---|
| Exact Diagonalization (Classical) | Small molecule ground states | Exponential in electron number | Intractable beyond ~50 orbitals [2] |
| Density Functional Theory (Classical) | Molecular structures, properties | Polynomial (typically O(N³)) | Fails for strongly correlated electrons [1] |
| Coupled Cluster (Classical) | Reaction energies, spectroscopy | O(Nâ¶) to O(N¹â°) | Prohibitively expensive for large systems [1] |
| Variational Quantum Eigensolver (Quantum) | Molecular ground states, reaction paths | Polynomial quantum + classical overhead | Current noise limits qubit count/accuracy [3] [2] |
| Quantum Phase Estimation (Quantum) | Precise energy calculations, excited states | Polynomial with fault tolerance | Requires fault-tolerant qubits [1] |
The exponential scaling of exact classical methods becomes apparent when modeling specific chemical systems. For instance, simulating the iron-molybdenum cofactor (FeMoco) essential for nitrogen fixation was estimated to require approximately 2.7 million physical qubits on a quantum computer, reflecting the immense complexity that makes this problem classically intractable [1]. Similarly, cytochrome P450 enzymes central to drug metabolism present similar computational challenges that exceed the capabilities of classical approximation methods [1].
Experimental Protocols & Performance Data
Hybrid Quantum-Classical Supercomputing for Complex Molecular Systems
Experimental Protocol: A collaborative team from Caltech, IBM, and RIKEN developed a quantum-centric supercomputing approach to study the [4Fe-4S] molecular cluster, a complex quantum system fundamental to biological processes including nitrogen fixation [4]. Their methodology proceeded as follows:
- Problem Encoding: The electronic structure problem of the [4Fe-4S] cluster was mapped onto a quantum computer using up to 77 qubits of an IBM Heron quantum processor [4].
- Quantum-Guided Reduction: Instead of using classical heuristics to approximate the Hamiltonian matrix (which grows exponentially with system size), the quantum computer identified the most crucial components of this matrix [4].
- Classical Refinement: The reduced Hamiltonian, containing only the most significant elements as determined by the quantum processor, was transferred to the RIKEN Fugaku supercomputer for exact diagonalization and computation of the final wave function and energy levels [4].
Performance Data: This hybrid approach successfully computed the electronic energy levels of the [4Fe-4S] cluster, a system that has long been a benchmark target for demonstrating quantum advantage in chemistry. The research did not definitively surpass all classical methods but significantly advanced the state-of-the-art in applying quantum algorithms to problems of real chemical interest [4].
Large-Scale Quantum Simulation Using FAST-VQE
Experimental Protocol: Kvantify, in partnership with IQM, implemented the FAST Variational Quantum Eigensolver (FAST-VQE) algorithm on a 50-qubit IQM Emerald quantum processor to study the dissociation curve of butyronitrile [2]. The methodology featured:
- Algorithm Selection: FAST-VQE was chosen over other VQE variants like ADAPT-VQE because it maintains a constant circuit count as the chemical system grows, enabling better scalability [2].
- Active Space Selection: The simulation utilized an active space of 50 molecular orbitals, a size that exceeds the practical limits of classical complete active space (CAS) methods [2].
- Hybrid Execution: Adaptive operator selection was performed on the quantum device, while energy estimation was handled by a chemistry-optimized classical simulator [2].
- Optimization Strategy: A greedy optimization strategy (adjusting one parameter at a time) was employed to overcome the bottleneck of simultaneous parameter optimization, allowing 120 iterations per hardware slot compared to just 30 with the full-parameter approach [2].
Performance Data: The 50-qubit implementation demonstrated measurable advantages over random baseline approaches, with the quantum hardware achieving faster convergence despite noise [2]. The greedy optimization strategy delivered an energy improvement of approximately 30 kcal/mol over the full-parameter optimization method [2]. This experiment highlighted a crucial shift in scaling limitations: as quantum hardware matures, classical optimization is becoming the primary bottleneck in hybrid algorithms [2].
Table 2: Performance Comparison of Recent Quantum Chemistry Experiments
| Experiment | Hardware Platform | Algorithm | System Studied | Key Performance Metric |
|---|---|---|---|---|
| Caltech/IBM/RIKEN [4] | IBM Heron (77 qubits) + Fugaku Supercomputer | Quantum-Centric Supercomputing | [4Fe-4S] molecular cluster | Successfully computed electronic energy levels of a previously intractable system |
| Kvantify/IQM [2] | IQM Emerald (50 qubits) | FAST-VQE | Butyronitrile dissociation | Achieved ~30 kcal/mol energy improvement with greedy optimization |
| IonQ Collaboration [5] | IonQ Forte | Quantum-Classical AFQMC | Carbon capture materials | Accurately computed atomic-level forces beyond classical accuracy |
| Google Quantum AI [6] | Willow processor | Quantum Echoes (OTOC) | Molecular structures via NMR | 13,000x speedup vs. fastest classical supercomputers |
Visualization of Workflows and Scaling Relationships
Computational Scaling Pathways
Figure 1: This diagram contrasts how classical and quantum computing resources scale with increasing chemical problem complexity. Classical methods face exponential resource growth, while quantum computing offers polynomial scaling.
Hybrid Quantum-Classical Workflow
Figure 2: The hybrid workflow used in modern quantum chemistry experiments, showing the iterative interaction between quantum and classical computing resources.
Table 3: Key Resources for Quantum Computational Chemistry Research
| Resource Category | Specific Examples | Function & Application |
|---|---|---|
| Quantum Software Development Kits | Qiskit (IBM) [7] [8], Cirq (Google) [3], Qrunch (Kvantify) [2] | Provide tools for building, optimizing, and executing quantum circuits; enable algorithm development and resource management |
| Quantum Hardware Platforms | IBM Heron/Nighthawk [7], IQM Emerald [2], IonQ Forte [5] | Physical quantum processors for running chemical simulations; vary in qubit count, connectivity, and error rates |
| Quantum Algorithms | Variational Quantum Eigensolver (VQE) [3], Quantum Approximate Optimization (QAOA) [3], Quantum-Classical AFQMC [5] | Specialized protocols for solving specific chemistry problems like ground state energy calculation and force estimation |
| Classical Co-Processors | High-Performance Computing clusters [7] [4], GPU accelerators | Handle computationally intensive classical components of hybrid algorithms, including error mitigation and parameter optimization |
| Error Mitigation Tools | Dynamic circuits [7], HPC-powered error mitigation [7], Zero-noise extrapolation | Improve result accuracy by suppressing and correcting for quantum processor noise and decoherence |
The experimental evidence demonstrates that quantum computing is progressively overcoming the exponential scaling problems that limit classical computational methods in chemistry. While today's quantum devices still face significant challenges in qubit count, connectivity, and error rates, the hybrid quantum-classical approaches demonstrated by leading research groups enable researchers to explore chemically relevant problems that were previously intractable [4] [2].
The field is rapidly advancing, with IBM projecting quantum advantage by the end of 2026 and fault-tolerant quantum computing by 2029 [7]. For researchers in chemistry and drug development, these developments signal a coming transformation in how molecular systems are simulated and understood. The ongoing collaboration between quantum hardware engineers, algorithm developers, and chemistry domain experts remains essential to fully realize the potential of quantum computing to solve chemistry's most challenging problems [9].
Limitations of Density Functional Theory (DFT) for Strongly Correlated Electrons
In the landscape of computational chemistry and materials science, the fundamental challenge revolves around the quantum mechanical many-body problem, whose computational complexity scales exponentially with system size. Density Functional Theory (DFT) has emerged as the cornerstone electronic structure method for quantum simulations across chemistry, physics, and materials science due to its favorable balance between accuracy and computational cost, typically scaling as O(N³) with system size. However, this favorable scaling comes at a significant cost: the method's accuracy is fundamentally limited by approximations in the exchange-correlation functional, a limitation that becomes critically pronounced in strongly correlated electron systems. These systems, characterized by competing quantum interactions that prevent electrons from moving independently, exhibit some of the most intriguing phenomena in condensed matter physics, including high-temperature superconductivity, colossal magnetoresistance, and metal-insulator transitions [10].
The core challenge lies in the failure of standard DFT functionals (LDA, GGA) to adequately capture the strong electron-electron interactions in these materials. While DFT succeeds tremendously for weakly correlated systems, its approximations fundamentally break down when electron localization and dynamic correlations dominate the physical behavior. This limitation has profound implications for drug development professionals and chemical researchers studying transition metal complexes, catalytic reaction centers, and quantum materials, where predictive accuracy is essential for rational design. This review systematically examines the theoretical origins, practical manifestations, and computational solutions for DFT's limitations in strongly correlated systems, providing researchers with a comprehensive framework for selecting appropriate methodologies beyond conventional DFT.
Fundamental Limitations of Standard DFT Approximations
The Self-Interaction Error and Electronic Delocalization
The foundational issue plaguing conventional DFT approximations is the self-interaction error (SIE), where an electron incorrectly interacts with itself. In exact DFT, this spurious self-interaction would precisely cancel, but approximate functionals fail to achieve this cancellation, leading to unphysical delocalization of electronic states. This error profoundly impacts predicted material properties, as evidenced in studies of europium hexaboride (EuB₆) where standard functionals fail to capture subtle symmetry breaking under pressure [11]. The SIE becomes particularly detrimental in strongly correlated materials containing localized d- and f-electrons, where electronic states should remain spatially confined due to strong Coulomb repulsion.
Standard DFT approximations tend to underestimate band gaps and predict metallic behavior for systems that are experimentally observed to be insulators or semiconductors. This failure stems from the inherent difficulty in describing static correlation effects, where multiple electronic configurations contribute significantly to the ground state. The delocalization tendency of conventional functionals presents a critical limitation for drug development researchers studying transition metal-containing enzymes or investigating charge transfer processes in photopharmaceuticals, where accurate prediction of electronic structure is prerequisite for understanding mechanism.
Limitations of DFT+U and Hybrid Functional Approaches
Two predominant strategies have emerged to address these limitations:
DFT+U Approach: This method introduces an effective on-site Coulomb interaction parameter (U) to localize electrons and correct the self-interaction error for specific orbitals [10]. While DFT+U can improve descriptions of localized states, it introduces empirical parameters whose determination often requires experimental calibration, limiting its predictive power. The approach has shown promise in systems like EuB₆ when combined with meta-GGA functionals exhibiting reduced SIE [11].
Hybrid Functionals: These methods incorporate a fraction of exact Hartree-Fock exchange with DFT exchange-correlation, partially mitigating the self-interaction error [10]. While offering improved accuracy for many molecular systems, hybrid functionals face significant challenges for strongly correlated solids, where the appropriate mixing parameter is difficult to determine a priori and computational cost increases substantially.
Table 1: Comparison of Standard DFT Approaches for Strongly Correlated Systems
| Method | Key Principle | Advantages | Limitations for Strongly Correlated Systems |
|---|---|---|---|
| LDA/GGA | Local density approximation/generalized gradient approximation | Computational efficiency; Good for weakly correlated systems | Severe self-interaction error; Underestimates band gaps; Favors metallic states |
| DFT+U | Adds Hubbard U parameter to localize electrons | Corrects delocalization error for specific orbitals; Improved band gaps | U parameter often empirical; Requires experimental calibration; Not fully first-principles |
| Hybrid Functionals | Mixes Hartree-Fock exchange with DFT exchange-correlation | Reduces self-interaction error; Improved molecular properties | High computational cost; Optimal mixing parameter difficult to determine for solids |
Advanced Methodologies Beyond Conventional DFT
Embedding Approaches: Combining DFT with Many-Body Theories
Sophisticated embedding methodologies have emerged that combine the computational efficiency of DFT with accurate many-body theories for treating strongly correlated subspaces:
DFT+DMFT (Dynamical Mean Field Theory): This approach maps the bulk quantum many-body problem onto an impurity model coupled to a self-consistent bath, capturing local temporal fluctuations absent in conventional DFT [10]. DFT+DMFT successfully describes aspects of the electronic structure of correlated materials, but challenges remain in capturing non-local spin fluctuations and vertex corrections beyond the random phase approximation.
Tensor Network Methods: Recent breakthroughs have demonstrated the powerful combination of DFT with tensor networks, particularly for one-dimensional and quasi-one-dimensional materials [10] [12]. This approach uses DFT with the constrained random phase approximation (cRPA) to construct an effective multi-band Hubbard model, which is then solved using matrix product states (MPS). The method provides systematic control over accuracy through the bond dimension and scales efficiently with system size, enabling quantitative prediction of band gaps, spin magnetization, and excitation energies.
The Strong-Interaction Limit of DFT
The strictly correlated electrons (SCE) functional represents the strong-interaction limit of DFT and provides a formally exact approach for addressing strong correlation [13]. This framework reformulates DFT as an optimal transport problem with Coulomb cost, offering insights into the exact form of the exchange-correlation functional in the strong-correlation regime. Integration of the SCE approach into the Kohn-Sham framework (KS-SCE) has shown promising results, such as correctly dissociating Hâ‚‚ molecules where standard approximations fail [13].
Table 2: Advanced Computational Methods for Strongly Correlated Systems
| Method | Theoretical Foundation | System Dimensionality | Key Observables | Computational Scaling |
|---|---|---|---|---|
| DFT+DMFT | Dynamical mean field theory; Quantum impurity models | 3D bulk systems | Spectral functions; Metal-insulator transitions | O(N³) to O(Nâ´) with high prefactor |
| Tensor Networks (MPS) | Matrix product states; Renormalization group | 1D and quasi-1D systems | Band gaps; Spin magnetization; Excitation energies | Efficient with system size; Tunable via bond dimension |
| SCE-DFT | Strictly correlated electrons; Optimal transport theory | Molecular systems | Strong-interaction limit; Dissociation curves | Varies with implementation |
Research Toolkit for Strongly Correlated Systems
Essential Computational Reagents and Methodologies
Researchers investigating strongly correlated materials require specialized computational tools to overcome DFT limitations:
cRPA (Constrained Random Phase Approximation): A downfolding technique for constructing effective low-energy models by integrating out high-energy degrees of freedom while calculating screened interaction parameters [10] [12].
Multi-band Hubbard Models: Effective Hamiltonians containing essential physics of correlated materials, with parameters derived from first-principles calculations [10].
Tensor Network Solvers: Mathematical engines based on matrix product states (MPS) and projected entangled-pair states (PEPS) that efficiently represent quantum many-body wavefunctions [10].
Advanced Exchange-Correlation Functionals: Meta-GGAs and double-hybrid functionals with reduced self-interaction error for improved treatment of correlated materials [11].
Experimental Validation Protocols
Accurate assessment of computational methodologies requires comparison with experimental observables:
Band Gap Measurements: Direct comparison between computed and experimentally determined band gaps provides a crucial validation metric [10] [12].
Angle-Resolved Photoemission Spectroscopy (ARPES): Directly probes electronic band structure and many-body effects such as spin-charge separation [10].
X-ray Absorption Near Edge Structure (XANES): Provides element-specific information about electronic states and local symmetry, as employed in EuB₆ studies under pressure [11].
Workflow for Advanced Strong Correlation Analysis
The following diagram illustrates the integrated computational workflow for treating strongly correlated materials, combining first-principles calculations with many-body theories:
Computational Workflow for Correlated Materials
This workflow demonstrates the multi-scale approach required for quantitative descriptions of strongly correlated materials, beginning with conventional DFT calculations and progressing through model construction to advanced many-body solutions.
The limitations of standard DFT for strongly correlated electrons represent a fundamental challenge at the heart of computational quantum chemistry and materials science. While conventional DFT approaches provide an essential starting point with favorable computational scaling, their failures in predicting electronic properties of correlated materials necessitate advanced methodologies that explicitly treat many-body effects. The integration of DFT with many-body theories such as tensor networks, dynamical mean field theory, and the strictly correlated electrons framework represents the frontier of computational research for strongly correlated systems.
For researchers in drug development and chemical design, these advances offer potential pathways to accurate simulation of transition metal catalysts, photopharmaceutical mechanisms, and electronic processes in complex molecular systems. The ongoing development of systematically improvable, computationally efficient methods that bridge quantum and classical scaling paradigms will continue to enhance our fundamental understanding and predictive capabilities for the most challenging strongly correlated materials.
The claim of discovering a room-temperature superconductor, LK-99, sent shockwaves through the scientific community in 2023. This material, a copper-doped lead-oxyapatite (Pb({9})CuP({6})O(_{25})), was purported to exhibit superconductivity at temperatures as high as 400 K (127 °C) under ambient pressure [14]. Such a discovery promised to revolutionize technologies from energy transmission to quantum computing. However, the subsequent global effort to replicate these results unveiled a more sobering reality: profound gaps in our fundamental knowledge and methodologies, particularly in the interplay between classical computational prediction and experimental validation in materials science. This case study examines the LK-99 saga, comparing the performance of theoretical and experimental "protocols" and framing the findings within the broader thesis of quantum versus classical computational scaling in chemistry research.
Experimental Replication: A Consensus of Negative Results
Despite initial global excitement, the consensus that emerged from numerous independent replication attempts was that LK-99 is not a room-temperature superconductor [14]. The following table summarizes key experimental results from peer-reviewed studies and reputable replication efforts, which collectively failed to observe the definitive signatures of superconductivity.
Table 1: Summary of Key Experimental Replication Attempts on LK-99
| Research Group / Study | Synthesis & Methodology Highlights | Key Experimental Results | Conclusion on Superconductivity |
|---|---|---|---|
| Cho et al. (2024) [15] | Synthesized LK-99 under various cooling conditions; used Powder X-ray Diffraction (PXRD) for phase analysis. | Slow cooling increased LK-99 phase but also retained impurities. No Meissner effect observed at ambient temperature or in liquid nitrogen. High electrical resistance. | Absence of superconductivity confirmed. Magnetic responses attributed to ferromagnetic impurities. |
| K. Kumar et al. (2023) [16] | Synthesized sample at 925°C; standard protocol from original preprints. | No large-area superconductivity observed at room temperature. No magnetic levitation (Meissner effect) detected. | No evidence of superconductivity in the synthesized sample. |
| PMC Study (2023) [17] | Used high-purity precursors; rigorous phase verification via PXRD and Rietveld refinement. Four-probe resistivity measurement. | Sample was highly resistive, showing insulator-like behavior from 215 to 325 K. Magnetization measurements indicated diamagnetism, not superconductivity. | Confirmed absence of superconductivity in phase-pure LK-99. |
| Beijing University Study [16] | Reproduced the synthesis process precisely. | Synthesized material placed on a magnet produced no repulsion and no magnetic levitation was observed. | No support for the room-temperature superconductor claim. |
| Leslie Schoop (Princeton) [18] | Simple replication attempt; visual and basic property checks. | Resulting crystals were transparent, unlike the opaque material in original claims, indicating different composition/impurities. | LK-99 is not a superconductor. |
| (2S)-2,6-Diamino-2-methylhexanoic acid | (2S)-2,6-Diamino-2-methylhexanoic Acid | Bench Chemicals | |
| 10-Acetoxy-8,9-epoxythymol isobutyrate | 10-Acetoxy-8,9-epoxythymol isobutyrate|High-Quality Reference Standard | 10-Acetoxy-8,9-epoxythymol isobutyrate (CAS 106009-86-3) is for research applications such as antimicrobial and cytotoxicity studies. For Research Use Only. Not for human or veterinary use. | Bench Chemicals |
The most definitive experiments measured the material's electrical transport properties, consistently finding that LK-99 is a highly resistive insulator, not a zero-resistance superconductor [17]. The occasional observations of partial magnetic levitation, initially misinterpreted as the Meissner effect, were later attributed to ferromagnetic or diamagnetic impurities like copper(I) sulfide (Cu(_{2})S) that form during the synthesis [14] [19].
The Computational Divide: Classical Predictions vs. Quantum Reality
The LK-99 episode highlighted a critical vulnerability in modern materials research: the over-reliance on and potential misinterpretation of classical computational methods.
The Role of Density Functional Theory (DFT) and Its Shortcomings
Classical computational methods, particularly Density Functional Theory (DFT), were rapidly deployed to assess LK-99's viability. Shortly after the initial claim, a study from Lawrence Berkeley National Laboratory used DFT to analyze LK-99 and suggested its structure might host isolated flat bands that could contribute to superconductivity [16] [14]. This theoretical finding was initially seized upon as validation.
However, this optimism exposed a key limitation. DFT, while powerful, operates within the framework of classical computing and has significant shortcomings when modeling complex quantum systems. As solid-state chemist Professor Leslie Schoop pointed out, a major flaw was that these early DFT calculations assumed the crystal structure proposed in the original, unverified preprint [18]. The adage "garbage in, garbage out" applies; an incorrect initial structure guarantees an incorrect electronic structure prediction. Furthermore, standard DFT methods often struggle with strongly correlated electron systems, precisely the kind of physics that might underpin high-temperature superconductivity.
The Quantum Computing Promise
This is where the potential of quantum computing becomes apparent. Unlike classical computers that use bits (0 or 1), quantum computers use qubits, which can exist in superpositions of 0 and 1 simultaneously [20]. This property of "massive quantum parallelism" allows them to naturally simulate quantum mechanical systems [21].
For a problem like predicting a new superconductor, a fault-tolerant quantum computer could, in theory, directly and accurately simulate the many-body quantum interactions within a material's crystal structure. This would circumvent the approximations required by DFT and provide a more reliable prediction of properties like superconductivity before costly and time-consuming experimental synthesis is undertaken. The scaling is fundamentally different: while classical computing power for such simulations grows linearly or polynomially with system complexity, effectively managed quantum computational power could grow exponentially for these specific tasks [20] [22].
Table 2: Classical vs. Quantum Computing in Materials Simulation
| Feature | Classical Computing (e.g., DFT) | Quantum Computing (Potential) |
|---|---|---|
| Basic Unit | Bit (0 or 1) | Qubit (0, 1, or both) |
| Underlying Principle | Binary Logic | Quantum Mechanics (Superposition, Entanglement) |
| Approach to Electron Correlation | Uses approximate functionals; can fail with strong correlations | Naturally handles entanglement and superposition |
| Scaling for Quantum Simulations | Polynomial to exponential, leading to intractable calculations | Theoretically polynomial for exact simulation |
| Maturity for Materials Science | Mature, widely used, but with known limitations | Emerging; requires fault-tolerant hardware not yet available |
| Outcome in LK-99 Case | Provided conflicting and ultimately misleading signals | Could have provided a more definitive theoretical assessment |
The Scientist's Toolkit: Essential Reagents and Methods for LK-99 Research
The synthesis and analysis of LK-99 require specific precursors and sophisticated instrumentation. The following table details the key research reagents and their functions in the typical experimental protocol.
Table 3: Key Research Reagent Solutions for LK-99 Synthesis and Analysis
| Reagent / Material | Function in the Experiment | Key Characteristics & Notes |
|---|---|---|
| Lead(II) Oxide (PbO) | Precursor for synthesizing Lanarkite (Pbâ‚‚SOâ‚…). | High-purity powder is essential to minimize impurities. |
| Lead(II) Sulfate (PbSOâ‚„) | Co-precursor for synthesizing Lanarkite (Pbâ‚‚SOâ‚…). | Freshly prepared and dried to ensure phase purity [17]. |
| Copper (Cu) Powder | Precursor for synthesizing Copper(I) Phosphide (Cu₃P). | High purity (e.g., 99.999%); checked for absence of CuO [17]. |
| Phosphorus (P) Grains | Precursor for synthesizing Copper(I) Phosphide (Cu₃P). | Handling in inert atmosphere (e.g., Argon glovebox) is critical due to reactivity [15]. |
| Copper(I) Phosphide (Cu₃P) | Final precursor reacted with Lanarkite to produce LK-99. | Phase purity is crucial; unreacted copper can lead to impurities [17]. |
| Lanarkite (Pb₂SO₅) | Final precursor mixed with Cu₃P to produce LK-99. | Synthesized by heating PbO and PbSO₄ at 725°C for 24 hours [14]. |
| Quartz Tube/Ampoule | Reaction vessel for synthesis steps. | Must withstand high temperatures (up to 925°C) and vacuum (10â»Â² to 10â»âµ Torr) [15] [17]. |
| Powder X-ray Diffractometer (PXRD) | Primary tool for verifying the crystal structure and phase purity of all precursors and the final product. | Data is analyzed with Rietveld refinement software (e.g., FullProf) for quantitative phase analysis [15] [17]. |
| Physical Property Measurement System (PPMS) | Measures electrical transport properties (e.g., resistivity) under varying temperatures and magnetic fields. | Used in a four-probe configuration to accurately measure the resistance of the sample [17]. |
| SQUID Magnetometer | Measures the magnetic properties of a material with high sensitivity. | Used to detect diamagnetism and check for the Meissner effect, a hallmark of superconductivity [17]. |
| 1-Cyanoethyl(diethylamino)dimethylsilane | 1-Cyanoethyl(diethylamino)dimethylsilane | 1-Cyanoethyl(diethylamino)dimethylsilane is a silane reagent for surface modification, polymer synthesis, and thin film deposition. For Research Use Only. Not for human use. |
| 3-Iodo-N-[(benzyloxy)carbonyl]-L-tyrosine | 3-Iodo-N-[(benzyloxy)carbonyl]-L-tyrosine, CAS:79677-62-6, MF:C17H16INO5, MW:441.22 g/mol | Chemical Reagent |
Detailed Experimental Protocol: Synthesizing and Testing LK-99
The following diagram illustrates the comprehensive multi-step workflow for synthesizing and characterizing LK-99, integrating the reagents and methods from the toolkit.
Diagram Title: LK-99 Synthesis and Characterization Workflow
Synthesis Protocol:
Precursor Preparation:
- Synthesis of Lanarkite (Pb₂SO₅): Mix lead(II) oxide (PbO) and lead(II) sulfate (PbSO₄) powders in a 1:1 molar ratio. Pelletize the mixture and heat it in an alumina crucible at 725 °C for 24 hours [17]. The product is a white solid.
- Synthesis of Copper(I) Phosphide (Cu₃P): In an argon-filled glovebox to prevent oxidation, uniformly grind together copper powder and phosphorus grains in a 3:1 molar ratio [15]. Pelletize the mixture, seal it in an evacuated (10â»Â² to 10â»âµ Torr) quartz tube, and heat it at 550 °C for 48 hours [14] [17].
Final LK-99 Synthesis: Thoroughly grind the synthesized Lanarkite and Copper(I) Phosphide crystals together in a stoichiometric ratio. Pelletize the mixed powder, seal it in an evacuated quartz tube, and react it at a high temperature of 925 °C for 5 to 20 hours [15] [14]. The resulting product is a gray-black, polycrystalline solid.
Characterization Protocol:
- Structural Analysis (PXRD): Crush the final product into a fine powder and analyze it using Powder X-ray Diffraction (PXRD). The data should be refined using the Rietveld method (e.g., with FullProf software) to confirm the formation of the lead-apatite crystal structure and quantify the presence of any impurity phases, such as Cuâ‚‚S [15] [17].
- Electrical Transport Measurement: Use a four-probe resistivity measurement setup within a Physical Property Measurement System (PPMS). This method eliminates the contribution of contact resistance, allowing for accurate measurement of the sample's intrinsic resistance as a function of temperature (e.g., from 215 K to 325 K) [17].
- Magnetic Property Measurement: Use a SQUID (Superconducting Quantum Interference Device) magnetometer to measure the sample's magnetization with high sensitivity. This test looks for the definitive Meissner effect (perfect diamagnetism) and measures the magnetic susceptibility [17].
- Magnetic Levitation Test: Visually test small sample fragments by placing them on a permanent magnet (e.g., Ndâ‚‚Feâ‚â‚„B) at room temperature. A true superconductor would demonstrate stable levitation and flux pinning, not just a partial tilt due to ferromagnetism [16] [14].
The LK-99 story is not a tale of failure but a powerful case study in the scientific process. It underscores a critical gap in our current research paradigm: the limitations of classical computational methods in predicting and explaining complex quantum phenomena in materials. While DFT is an invaluable tool, its misapplication in the absence of robust experimental structures can lead the community down unproductive paths.
The path forward requires a more integrated and humble approach. Experimental synthesis must be performed with scrupulous attention to detail and phase purity, and theoretical predictions must be treated as guides rather than gospel. Ultimately, bridging this fundamental knowledge gap may hinge on the next computational revolution: the advent of practical quantum computing. By providing a native platform for simulating quantum matter, quantum computers could one day transform the search for revolutionary materials like room-temperature superconductors from a process of serendipitous discovery into one of principled design.
Molecular systems are, at their fundamental level, governed by the laws of quantum mechanics. The behavior of electrons and atomic nuclei involves quantum phenomena such as superposition, entanglement, and tunneling—effects that classical computers can simulate only with exponential resource growth. This inherent quantum nature makes molecular systems a putative native application for quantum processors (QPUs), which operate on the same physical principles [23] [24]. For computational chemistry, this suggests the potential for a profound advantage: quantum computers could simulate molecular processes with natural efficiency, potentially bypassing the steep approximations and computational costs that challenge even the most powerful classical supercomputers [25] [26].
The central challenge in classical computational chemistry is the exponential scaling of exact methods like Full Configuration Interaction (FCI) with system size. While approximate methods like Density Functional Theory (DFT) or Coupled Cluster offer more favorable scaling, they can fail for systems with strong electron correlation, such as transition metal catalysts or complex biomolecules [25] [26]. Quantum algorithms, particularly Quantum Phase Estimation (QPE), offer a promising alternative with the potential for polynomial scaling for these problems [26]. This guide provides an objective comparison of the current performance landscape between classical and quantum computational chemistry approaches, detailing the experimental protocols and hardware requirements that underpin recent advancements.
Computational Scaling: A Theoretical and Practical Comparison
The theoretical advantage of quantum computing in chemistry stems from the different ways classical and quantum algorithms scale with problem size, typically measured by the number of basis functions (N). The table below summarizes the expected timelines for quantum algorithms to surpass various classical methods for a representative high-accuracy target.
Table 1: Projected timelines for quantum phase estimation (QPE) to surpass classical computational chemistry methods for a representative high-accuracy target (error < 1mHa). Adapted from [26].
| Computational Method | Classical Time Complexity | Projected Year for QPE Surpassment |
|---|---|---|
| Density Functional Theory (DFT) | O(N³) | Beyond 2050 |
| Hartree-Fock (HF) | O(Nâ´) | Beyond 2050 |
| Møller-Plesset Second Order (MP2) | O(Nâµ) | Beyond 2050 |
| Coupled Cluster Singles & Doubles (CCSD) | O(Nâ¶) | ~2044 |
| CCSD with Perturbative Triples (CCSD(T)) | O(Nâ·) | ~2036 |
| Full Configuration Interaction (FCI) | O*(4^N) | ~2031 |
This analysis suggests that quantum computing will first disrupt the most accurate, classically intractable methods before competing with faster, less accurate approximations [26]. The polynomial scaling of QPE (O(N²/ϵ) for a target error ϵ) is expected to eventually overtake the exponential scaling of FCI and the high-order polynomial scaling of "gold standard" methods like CCSD(T). However, for the foreseeable future, low-accuracy methods like DFT will remain solidly in the classical computing domain [26].
Experimental Protocols in Hybrid Quantum-Classical Chemistry
Current quantum hardware, termed Noisy Intermediate-Scale Quantum (NISQ), is not yet capable of running long, fault-tolerant algorithms like QPE. Therefore, today's experimental focus is on hybrid quantum-classical algorithms that delegate the most quantum-native subproblems to the QPU while leveraging classical high-performance computing (HPC) for the rest [27] [9] [28].
The Quantum-Centric Supercomputing Approach (Caltech/IBM/RIKEN)
A leading protocol demonstrated in 2025 for studying the [4Fe-4S] molecular cluster—a complex iron-sulfur system relevant to nitrogen fixation—showcases this hybrid paradigm [27].
Objective: To determine the ground-state energy of the [4Fe-4S] cluster by solving the electronic Schrödinger equation. Classical Challenge: The Hamiltonian matrix for this system is too large to handle exactly. Classical heuristics prune this matrix, but their approximations can be unreliable [27]. Quantum Role: An IBM Heron quantum processor (using up to 77 qubits) was used to rigorously identify the most important components of the Hamiltonian matrix, replacing classical heuristics [27]. Workflow: The quantum computer processed the full problem to output a compressed, relevant subset of the Hamiltonian. This reduced matrix was then passed to the Fugaku supercomputer for final diagonalization to obtain the exact wave function and energy [27].
This "quantum-centric supercomputing" approach demonstrates a practical division of labor, using the QPU as a specialized accelerator for the most quantum-native task: identifying the essential structure of a complex quantum state [27].
The DMET-SQD Approach (Cleveland Clinic/IBM/Michigan State)
Another advanced protocol, the Density Matrix Embedding Theory with Sample-Based Quantum Diagonalization (DMET-SQD), was used to simulate molecular conformers of cyclohexane, a standard test in organic chemistry [28].
Objective: To compute the relative energies of different cyclohexane conformers with chemical accuracy (within 1 kcal/mol). Classical Challenge: Simulating entire large molecules exactly is infeasible; mean-field approximations ignore crucial electron correlations [28]. Quantum Role: The DMET method breaks the molecule into smaller fragments. The SQD algorithm, run on an IBM Eagle processor (using 27-32 qubits), simulated the quantum chemistry of these individual fragments. SQD is notably tolerant of the noise present in current-generation hardware [28]. Workflow: The global molecule is partitioned into fragments. A classical computer handles the bulk environment, while the quantum computer solves the embedded fragment problem via SQD. The results are integrated back classically to reconstruct the total energy [28]. Result: The hybrid DMET-SQD method achieved energy differences within 1 kcal/mol of classical benchmarks, validating its potential for biologically relevant molecules [28].
The following diagram visualizes the logical flow common to these hybrid computational workflows.
The Researcher's Toolkit for Hybrid Quantum Chemistry
Implementing the protocols above requires a suite of specialized hardware and software "reagents." The following table details the key components.
Table 2: Essential "Research Reagent Solutions" for current hybrid quantum-classical computational chemistry experiments.
| Tool Category | Specific Example | Function & Relevance |
|---|---|---|
| Quantum Hardware (QPU) | IBM Heron/Eagle Processors [27] [28] | Superconducting qubit processors that perform the core quantum computations; require milli-Kelvin cooling. |
| Classical HPC | Fugaku Supercomputer [27] | A world-class supercomputer that handles the computationally intensive classical portions of the hybrid algorithm. |
| Software Libraries | Qiskit [28] | An open-source SDK for working with quantum computers at the level of circuits, pulses, and algorithms. |
| Software Libraries | Tangelo [28] | An open-source quantum chemistry toolkit used to implement the DMET embedding framework. |
| Algorithmic Framework | Density Matrix Embedding Theory (DMET) [28] | A fragmentation technique that divides a large molecular problem into smaller, quantum-tractable fragments. |
| Algorithmic Framework | Sample-Based Quantum Diagonalization (SQD) [28] | A noise-resilient quantum algorithm used to solve for the energy of a quantum fragment on NISQ hardware. |
| Error Mitigation | Gate Twirling & Dynamical Decoupling [28] | Software-level techniques applied to quantum circuits to mitigate the effect of noise without full error correction. |
| Purine, 2,6-diamino-, sulfate, hydrate | Purine, 2,6-diamino-, sulfate, hydrate, CAS:116295-72-8, MF:C10H16N12O5S, MW:416.38 g/mol | Chemical Reagent |
| 2-(2-Amino-4-methoxyphenyl)acetonitrile | 2-(2-Amino-4-methoxyphenyl)acetonitrile | RUO | 2-(2-Amino-4-methoxyphenyl)acetonitrile, a key intermediate for heterocyclic synthesis. For Research Use Only. Not for human or veterinary use. |
Discussion and Future Horizons
The experimental data and protocols demonstrate that hybrid quantum-classical approaches are already yielding chemically meaningful results for small to medium-sized systems [27] [28]. The primary advantage of the QPU in these workflows is its ability to handle the strong electron correlations and exponential state spaces that challenge even the most powerful classical HPCs for certain problems [25] [26].
However, the path to a unambiguous "quantum advantage" in chemistry is still long. Current methods require heavy error mitigation and are limited by the number of reliable logical qubits. Experts estimate that robust fault-tolerant quantum computers capable of outperforming classical computers for high-accuracy problems like CCSD(T) or FCI are likely 10-20 years away [25] [26]. The field is actively pursuing a co-design strategy, where chemists, algorithm developers, and hardware engineers collaborate to identify the problems and refine the tools that will define the next decade of progress [9].
For researchers in drug development and materials science, the present utility of quantum computing lies in its role as a specialized accelerator within a larger HPC ecosystem. As hardware matures, its impact is projected to grow from highly accurate small-molecule simulations toward larger, more complex systems like enzymes and novel materials, fundamentally reshaping the landscape of computational discovery [25] [26] [9].
Quantum Algorithms in Action: From Theory to Real-World Chemistry Problems
In the field of computational chemistry and drug discovery, researchers face a fundamental challenge: the accurate simulation of molecular systems requires solving the Schrödinger equation, a task whose computational cost grows exponentially with system size on classical computers. Methods like Density Functional Theory (DFT) scale as ( \mathcal{O}(N^3) ), while more accurate approaches such as Coupled Cluster theory scale as steeply as ( \mathcal{O}(N^7) ), where ( N ) represents the number of electrons in the system [29]. This exponential scaling creates an insurmountable barrier for studying complex molecules relevant to pharmaceutical development, such as iron-sulfur clusters in enzymes or covalent drug-target interactions.
Quantum computing offers a potential pathway to overcome this bottleneck, as quantum systems can naturally simulate other quantum systems. However, current Noisy Intermediate-Scale Quantum (NISQ) hardware remains limited by qubit counts, connectivity constraints, and inherent noise. Hybrid Quantum-Classical (HQC) models have emerged as a strategic compromise, leveraging classical computers for the bulk of computational workload while delegating specific, quantum-native subroutines to quantum processors. This architecture creates a practical bridge to quantum advantage, enabling researchers to explore quantum algorithms on today's hardware while addressing real-world chemical problems [4] [29] [30].
Hybrid Model Architectures for Chemical Simulation
The Variational Quantum Eigensolver (VQE) Framework
The Variational Quantum Eigensolver (VQE) has become the cornerstone algorithm for quantum chemistry on NISQ devices. This hybrid approach combines a parameterized quantum circuit (PQC) with classical optimization to compute molecular properties, most commonly the ground state energy [30]. The quantum processor's role is to prepare and measure the quantum state of the molecular system, while the classical processor adjusts the circuit parameters to minimize the energy expectation value.
The VQE workflow follows these steps:
- Problem Mapping: The molecular Hamiltonian is transformed from fermionic to qubit representation using techniques like Jordan-Wigner or parity transformation.
- Ansatz Preparation: A parameterized quantum circuit (ansatz) is selected to prepare trial wavefunctions.
- Measurement and Optimization: The energy expectation value is measured on the quantum device, and a classical optimizer adjusts circuit parameters to minimize this energy.
This framework has been successfully applied to molecular systems of real-world relevance, including the study of prodrug activation mechanisms and covalent inhibitor interactions [30].
Quantum-Centric Supercomputing
A more recent architecture, termed "quantum-centric supercomputing," demonstrates how quantum and classical resources can be integrated at scale. In this approach, a quantum processor identifies the most critical components of large Hamiltonian matrices, which are then solved exactly on classical supercomputers. This division of labor was showcased in a landmark study where researchers used an IBM Heron quantum processor with up to 77 qubits to simplify the mathematics for an iron-sulfur molecular cluster, then leveraged the Fugaku supercomputer to solve the problem [4].
This methodology addresses a key bottleneck in quantum chemistry: classical algorithms often rely on approximations to prune down exponentially large Hamiltonian matrices. The quantum computer provides a more rigorous selection of relevant matrix components, potentially improving accuracy while reducing computational overhead [4].
Table: Hybrid Quantum-Classical Architectures for Chemical Simulation
| Architecture | Quantum Component Role | Classical Component Role | Key Applications |
|---|---|---|---|
| VQE [30] | State preparation and energy measurement | Parameter optimization and error mitigation | Molecular energy calculations, reaction profiling |
| Quantum-Centric Supercomputing [4] | Hamiltonian simplification and component selection | Large-scale matrix diagonalization | Complex molecular clusters, active space selection |
| Hybrid ML Potentials [29] | Feature embedding and non-linear transformation | Message passing and structural representation | Materials simulation, molecular dynamics |
Performance Comparison: Quantum-Classical Models vs. Classical Baselines
Chemical Accuracy in Molecular Simulations
Recent studies have provided quantitative comparisons between hybrid quantum-classical approaches and classical computational methods. In drug discovery applications, researchers have demonstrated that HQC models can achieve chemical accuracy while potentially reducing computational resource requirements for specific problem classes.
In one investigation focusing on prodrug activation—a critical process in pharmaceutical design—researchers computed Gibbs free energy profiles for carbon-carbon bond cleavage in β-lapachone derivatives. The hybrid quantum-classical pipeline employed a hardware-efficient ( R_y ) ansatz with a single layer as the parameterized quantum circuit for VQE. The results showed that the quantum computation agreed with Complete Active Space Configuration Interaction (CASCI) calculations, which serve as the reference exact solution within the active space approximation [30].
Table: Performance Comparison for Prodrug Activation Study [30]
| Computational Method | System Size (Qubits) | Accuracy vs. CASCI | Key Application |
|---|---|---|---|
| Classical DFT (M06-2X) | N/A | Reaction barrier consistent with experiment | C-C bond cleavage in β-lapachone |
| Classical CASCI | N/A | Reference method | Active space approximation |
| Hybrid VQE (R𑦠ansatz) | 2 | Consistent with CASCI | Quantum computation of reaction barrier |
Resource Efficiency and Scalability
Beyond accuracy metrics, hybrid models demonstrate advantages in resource efficiency. The application of HQC models to machine learning potentials (MLPs) for materials science reveals that replacing classical neural network components with variational quantum circuits can maintain accuracy while potentially reducing parameter counts. In benchmarks for liquid silicon simulations, hybrid quantum-classical MLPs achieved accurate reproduction of high-temperature structural and thermodynamic properties, matching classical state-of-the-art equivariant message-passing neural networks [29].
This efficiency stems from the ability of quantum circuits to generate highly complex non-linear transformations with relatively few parameters. The quantum processor executes targeted sub-tasks that supply additional expressivity, while the classical processor handles the bulk of computation [29]. This division of labor is particularly advantageous for NISQ devices, which remain constrained by qubit coherence times and gate fidelities.
Experimental Protocols for Hybrid Quantum-Classical Chemistry
Protocol 1: Quantum Computation of Reaction Profiles
The determination of Gibbs free energy profiles for chemical reactions represents a cornerstone application of quantum chemistry in drug discovery. The following protocol outlines the hybrid approach used to study covalent bond cleavage in prodrug activation [30]:
System Preparation:
- Select key molecules along the reaction coordinate
- Perform conformational optimization using classical methods
- Define active space (typically 2 electrons in 2 orbitals for C-C bond cleavage)
Hamiltonian Generation:
- Generate molecular Hamiltonian in fermionic form
- Apply parity transformation to convert to qubit representation
- Utilize the 6-311G(d,p) basis set for consistent comparison
Quantum Circuit Configuration:
- Implement hardware-efficient ( R_y ) ansatz with single layer
- Apply readout error mitigation techniques
- Execute on superconducting quantum processor (2 qubits)
Classical-VQE Integration:
- Use classical optimizer to minimize energy expectation value
- Employ polarizable continuum model (PCM) for solvation effects
- Calculate single-point energies with solvent corrections
Validation:
- Compare quantum results with classical CASCI and HF calculations
- Benchmark against experimental reaction feasibility
This protocol successfully demonstrated the computation of energy barriers for C-C bond cleavage, a crucial parameter in prodrug design that determines whether reactions proceed spontaneously under physiological conditions [30].
Protocol 2: Quantum-Centric Supercomputing for Complex Molecules
The study of the [4Fe-4S] molecular cluster—an important component in biological systems like the enzyme nitrogenase—required a more sophisticated protocol leveraging both quantum and classical resources at scale [4]:
Problem Decomposition:
- Define the full molecular system with all electrons and atomic positions
- Generate the complete Hamiltonian matrix
Quantum Pre-processing:
- Utilize IBM quantum device (Heron processor) with up to 77 qubits
- Identify the most important components in the Hamiltonian matrix
- Prune less relevant matrix elements using quantum measurements
Classical Post-processing:
- Feed the simplified Hamiltonian to Fugaku supercomputer
- Perform exact diagonalization on the reduced matrix
- Solve for the system's wave function and ground state energy
Validation and Analysis:
- Compare results with classical heuristic approaches
- Assess computational efficiency and accuracy gains
This protocol demonstrated that quantum computers can rigorously select relevant Hamiltonian components, potentially replacing the classical heuristics traditionally used for this task [4].
The Scientist's Toolkit: Essential Research Reagents
Implementing hybrid quantum-classical models requires specialized tools and frameworks that bridge the quantum-classical divide. The following table outlines key "research reagents" essential for conducting experiments in this domain:
Table: Essential Research Reagents for Hybrid Quantum-Classical Chemistry
| Tool/Platform | Type | Function | Example Use Case |
|---|---|---|---|
| TenCirChem [30] | Software Package | Quantum computational chemistry | VQE implementation for drug discovery |
| PyTorch/PennyLane [31] | Machine Learning Library | Hybrid model development | Physics-informed neural networks |
| OpenQASM [32] | Quantum Assembly Language | Quantum circuit representation | Benchmarking quantum algorithms |
| Hardware-Efficient Ansatz [30] | Quantum Circuit | State preparation | R𑦠ansatz for molecular simulations |
| RIKEN Fugaku [4] | Classical Supercomputer | Large-scale matrix diagonalization | Quantum-centric supercomputing |
| IBM Heron Processor [4] | Quantum Hardware | Quantum computation | 77-qubit chemical simulations |
| Oxacyclohexadec-12-en-2-one, (12E)- | Oxacyclohexadec-12-en-2-one, (12E)-, CAS:111879-80-2, MF:C15H26O2, MW:238.37 g/mol | Chemical Reagent | Bench Chemicals |
| 4-Decyltetradecan-1-ol | 4-Decyltetradecan-1-ol | High-Purity Long-Chain Fatty Alcohol | 4-Decyltetradecan-1-ol, a high-purity C24 fatty alcohol for research on lipids & surfactants. For Research Use Only. Not for human or veterinary use. | Bench Chemicals |
Hybrid quantum-classical models represent a pragmatic and powerful bridge to quantum computational advantage in chemistry and drug discovery. Current evidence demonstrates that these models can already tackle real-world problems, from prodrug activation kinetics to complex molecular cluster simulations, with accuracy comparable to classical methods [4] [30]. While definitive quantum advantage across all chemical applications remains on the horizon, the architectural patterns established by HQC models provide a clear pathway forward.
The strategic division of labor—where quantum processors handle naturally quantum subroutines while classical computers manage optimization, error mitigation, and large-scale data processing—enables researchers to extract maximum value from current NISQ devices. As quantum hardware continues to improve in scale and fidelity, the balance within these hybrid architectures will likely shift toward greater quantum responsibility, potentially unlocking the exponential scaling advantages promised by quantum mechanics for molecular simulation.
Variational Quantum Eigensolver (VQE) for Calculating Molecular Ground-State Energy
The calculation of molecular ground-state energies is a fundamental challenge in chemistry and drug discovery. Classical computational methods, such as density functional theory (DFT) and post-Hartree-Fock approaches, provide valuable insights but often fall short when applied to large systems and strongly correlated electrons, or when high accuracy is required [33]. The complexity of solving the electronic Schrödinger equation scales exponentially with system size on classical computers, creating an intractable bottleneck for simulating complex molecules or materials [34].
Quantum computing represents a paradigm shift, leveraging the principles of quantum mechanics to process information in ways that classical computers cannot [33]. The Variational Quantum Eigensolver (VQE) has emerged as a leading hybrid algorithm for the Noisy Intermediate-Scale Quantum (NISQ) era, offering a potential pathway to overcome classical scaling limitations [35]. This guide provides an objective comparison of VQE performance against classical alternatives, detailing experimental methodologies and presenting quantitative benchmarking data to inform researchers and drug development professionals.
Algorithmic Frameworks and Workflows
The VQE Algorithm: A Hybrid Approach
The Variational Quantum Eigensolver (VQE) is a hybrid quantum-classical algorithm that leverages the variational principle to approximate ground-state energies [35]. Fundamentally, VQE operates by:
- Parameterized Wavefunction: A trial wavefunction (ansatz) is prepared on a quantum processor using a parameterized quantum circuit, ( |\Psi(\boldsymbol{\theta})\rangle \equiv \hat{U}(\boldsymbol{\theta})|\Psi_0\rangle ).
- Expectation Measurement: The quantum computer measures the expectation value of the molecular Hamiltonian, ( E[\Psi(\boldsymbol{\theta})] = \langle \Psi(\boldsymbol{\theta}) | \hat{H} | \Psi(\boldsymbol{\theta}) \rangle ).
- Classical Optimization: A classical optimizer iteratively adjusts parameters ( \boldsymbol{\theta} ) to minimize the energy ( E[\Psi(\boldsymbol{\theta})] ), satisfying the variational principle ( E_g \leq E[\Psi(\boldsymbol{\theta})] ) [35].
This framework is particularly well-suited for NISQ devices because it uses quantum resources primarily for preparing and measuring quantum states, while offloading the optimization workload to classical computers [33].
Quantum-Classical Workflow
The following diagram illustrates the integrated workflow of a VQE calculation within a quantum-DFT embedding framework, as implemented in benchmarking studies [33]:
Advanced VQE Variants
Recent research has developed enhanced VQE variants to address limitations like barren plateaus and high measurement costs:
- ADAPT-VQE: Builds the ansatz iteratively, adding one quantum gate at a time based on the largest gradient. This gradient-driven strategy can bypass barren plateaus but is highly measurement-intensive [36].
- Greedy Gradient-Free Adaptive VQE (GGA-VQE): A "greedy" approach that selects operators and their optimal parameters in a single step. It uses only 2-5 circuit measurements per iteration and demonstrates superior noise resilience, having been successfully demonstrated on a 25-qubit quantum computer [36].
Experimental Protocols and Methodologies
Benchmarking Framework: The BenchQC Protocol
Recent systematic benchmarking studies, such as those using the BenchQC toolkit, have employed rigorous methodologies to evaluate VQE performance [33] [37] [38]:
- Molecular Systems: Focus on small aluminum clusters (Alâ», Alâ‚‚, Al₃â») chosen for intermediate complexity, relevance to materials science, and availability of reliable classical benchmarks from the Computational Chemistry Comparison and Benchmark DataBase (CCCBDB) [33].
- Quantum-DFT Embedding: The system is divided into a classical region (handled by DFT for core electrons) and a quantum region (handled by VQE for strongly correlated valence electrons) [33]. This hybrid approach mitigates current NISQ device limitations.
- Parameter Variation: Systematic testing of key parameters including:
- Classical optimizers (SLSQP, ADAM, BFGS, etc.)
- Circuit types (EfficientSU2, UCCSD, etc.)
- Basis sets (STO-3G, higher-level sets)
- Number of circuit repetitions
- Simulator types (statevector, noisy simulations)
- Reference Calculations: Results are compared against exact diagonalization using NumPy and reference data from CCCBDB to compute percent errors [33].
Classical Reference Methods
For context, VQE performance is typically compared against these classical computational chemistry methods:
- Full Configuration Interaction (FCI): Provides exact solutions within a given basis set but is computationally prohibitive for large systems.
- Coupled Cluster Theory (e.g., CCSD(T)): Considered the "gold standard" for quantum chemistry accuracy but scales steeply (O(Nâ·) for CCSD(T)).
- Density Functional Theory (DFT): More computationally efficient (O(N³)) but can be inaccurate for systems with strong electron correlation.
Performance Comparison and Benchmarking Data
VQE Configuration Performance
Comparative studies reveal how algorithmic choices significantly impact VQE performance. The table below summarizes key findings from benchmarking experiments on molecular systems:
Table 1: Performance of VQE Configurations on Molecular Systems
| Molecular System | Optimal Ansatz | Optimal Optimizer | Key Performance Metrics | Reference Method Error |
|---|---|---|---|---|
| Silicon atom [39] | UCCSD (with zero initialization) | ADAM | Most stable and precise results; close approximation to experimental values | N/A |
| Aluminum clusters (Alâ», Alâ‚‚, Al₃â») [33] | EfficientSU2 | SLSQP (among tested) | Percent errors consistently below 0.2% against CCCBDB | CCCBDB benchmarks |
| Hâ‚‚O, LiH [36] | GGA-VQE (adaptive) | Gradient-free greedy | Nearly 2x more accurate than ADAPT-VQE for Hâ‚‚O under noise; ~5x more accurate for LiH | Chemical accuracy threshold |
| 25-spin Ising model [36] | GGA-VQE (adaptive) | Gradient-free greedy | >98% fidelity on 25-qubit hardware; converged computation on NISQ device | Exact diagonalization |
Optimizer Performance Comparison
The choice of classical optimizer significantly impacts convergence efficiency and final energy accuracy:
Table 2: Classical Optimizer Performance in VQE Calculations
| Optimizer | Full Name | Convergence Efficiency | Stability | Computational Cost |
|---|---|---|---|---|
| SLSQP [33] | Sequential Least Squares Programming | Efficient convergence in benchmark studies | Stable for small molecules | Moderate |
| ADAM [39] | Adaptive Moment Estimation | Superior for silicon atom with UCCSD | Robust with zero initialization | Moderate |
| L-BFGS-B [40] | Limited-memory BFGS | Fast convergence when stable | Can get stuck in local minima | Low-memory, efficient |
| SPSA [40] | Simultaneous Perturbation Stochastic Approximation | Resilient to noise | Suitable for noisy hardware | Very low (few measurements) |
| AQGD [40] | Alternating Quantum Gradient Descent | Quantum-aware optimization | Moderate | Moderate |
| COBYLA [40] | Constrained Optimization By Linear Approximation | Gradient-free, reasonable convergence | Less efficient for high dimensions | Low |
Ansatz Performance Comparison
The ansatz choice balances expressibility against quantum resource requirements:
Table 3: Quantum Ansatz Comparison for Molecular Simulations
| Ansatz Type | Description | Strengths | Weaknesses | Hardware Efficiency |
|---|---|---|---|---|
| UCCSD [39] | Unitary Coupled Cluster Singles and Doubles | Chemically inspired, high accuracy for silicon atom | Deeper circuits, more gates | Low on current devices |
| EfficientSU2 [33] | Hardware-efficient parameterized circuit | Low-depth, tunable expressiveness | Does not conserve physical symmetries | High for NISQ devices |
| k-UpCCGSD [39] | Unitary Pair Coupled Cluster with Generalized Singles and Doubles | Moderate accuracy with reduced depth | Less accurate than UCCSD | Moderate |
| ParticleConservingU2 [39] | Particle-conserving universal 2-qubit ansatz | Remarkably robust across optimizers | May be less expressive | Moderate |
| GGA-VQE [36] | Greedy gradient-free adaptive ansatz | Noise-resilient, minimal measurements | Less flexible final circuit | Very high (2-5 measurements/iteration) |
The Scientist's Toolkit: Essential Research Reagents
Implementing VQE experiments requires both computational and chemical resources. The table below details key "research reagent" solutions for VQE experiments in computational chemistry:
Table 4: Essential Research Reagents and Computational Tools for VQE Experiments
| Tool/Category | Specific Examples | Function/Role | Implementation Notes |
|---|---|---|---|
| Quantum Software Platforms | Qiskit (v43.1) [33], CUDA-Q [41], InQuanto [41] | Provides interfaces for quantum algorithm implementation, circuit design, and execution | Qiskit Nature's ActiveSpaceTransformer used for orbital selection |
| Classical Computational Chemistry | PySCF [33], NumPy [33] | Performs initial orbital analysis; provides exact diagonalization benchmarks | Integrated within Qiskit framework for seamless workflow |
| Molecular Databases | CCCBDB [33], JARVIS-DFT [33] | Sources of pre-optimized molecular structures and benchmark data | Provides reliable ground-truth data for validation |
| Classical Optimizers | SLSQP, ADAM, L-BFGS-B, SPSA [40] | Adjusts quantum circuit parameters to minimize energy | Choice depends on convergence needs and noise resilience |
| Quantum Ansätze | UCCSD, EfficientSU2, Hardware-efficient [39] | Forms parameterized trial wavefunctions for VQE | Balance between chemical accuracy and NISQ feasibility |
| Error Mitigation Techniques | Zero-noise extrapolation, Probabilistic error cancellation [39] | Reduces impact of hardware noise without full error correction | Essential for obtaining meaningful results on real devices |
| Active Space Tools | ActiveSpaceTransformer (Qiskit Nature) [33] | Selects chemically relevant orbitals for quantum computation | Focuses computational resources on important regions |
| 1(or 2)-(2-Ethylhexyl) trimellitate | 1(or 2)-(2-Ethylhexyl) trimellitate | High-Purity RUO | 1(or 2)-(2-Ethylhexyl) trimellitate, a high-purity plasticizer & solvent for material science research. For Research Use Only. Not for human or veterinary use. | Bench Chemicals |
| N-(4-methylpyridin-2-yl)acetamide | N-(4-methylpyridin-2-yl)acetamide | Research Chemical | N-(4-methylpyridin-2-yl)acetamide for research applications. This product is For Research Use Only (RUO). Not for human or veterinary use. | Bench Chemicals |
The systematic benchmarking of VQE reveals a nuanced picture of its current capabilities and future potential for calculating molecular ground-state energies. When appropriately configured with optimal ansatzes, optimizers, and initialization strategies, VQE can achieve remarkable accuracy, with percent errors below 0.2% for small aluminum clusters compared to classical benchmarks [33]. The development of noise-resilient variants like GGA-VQE, which has been successfully demonstrated on a 25-qubit quantum computer, represents a significant step toward practical quantum advantage on NISQ devices [36].
However, substantial challenges remain. Quantum noise severely degrades VQE performance, necessitating robust error mitigation strategies [39]. The optimal configuration (ansatz, optimizer, initialization) appears to be system-dependent, requiring careful benchmarking for each new class of molecules [39]. While VQE shows promise for quantum chemistry applications, including drug discovery [34], its scalability to large, complex molecular systems awaits advances in both quantum hardware and algorithm design.
The quantum-classical hybrid approach of VQE, particularly when embedded within DFT frameworks, offers a pragmatic pathway for leveraging current quantum resources while mitigating their limitations. As quantum hardware continues to evolve, VQE and its variants may ultimately fulfill their potential to overcome the fundamental scaling limitations of classical computational chemistry methods.
The simulation of drug-target interactions represents a cornerstone of modern computational chemistry, essential for understanding mechanisms of drug action and designing new therapeutics. This challenge is particularly acute for high-impact targets like the KRAS oncogene, a key driver in pancreatic, colorectal, and lung cancers that has historically been considered "undruggable" due to its smooth surface and picomolar affinity for nucleotides [42] [43]. The central thesis in modern computational chemistry posits that quantum computing algorithms offer fundamentally superior scaling properties for simulating complex biochemical systems compared to classical computational approaches. As drug discovery confronts the vastness of chemical space (~10â¶â° molecules) and the complexity of biological systems, classical computing faces intrinsic limitations in processing power and algorithmic efficiency [44] [45]. This review objectively compares emerging quantum workflows against established classical methods for simulating KRAS inhibition, providing performance data, experimental protocols, and analytical frameworks to guide researchers in selecting appropriate computational strategies.
KRAS Biology and Therapeutic Significance
The Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) protein functions as a molecular switch, cycling between active GTP-bound and inactive GDP-bound states to regulate critical cellular signaling pathways including MAPK and PI3K-AKT [42]. Oncogenic mutations, most frequently at codons 12, 13, and 61, impair GTP hydrolysis and lock KRAS in a constitutively active state, driving uncontrolled cell proliferation and survival [43]. KRAS mutations demonstrate distinct tissue-specific prevalence patterns: G12D and G12V dominate in pancreatic ductal adenocarcinoma, G12C in lung adenocarcinoma (particularly among smokers), and A146 mutations primarily in colorectal cancer [42]. This mutational landscape creates a complex therapeutic targeting environment requiring sophisticated computational approaches.
Table 1: Prevalence of Major KRAS Mutations in Human Cancers
| Mutation | Primary Cancer Associations | Approximate Prevalence |
|---|---|---|
| G12D | Pancreatic, Colorectal | ~33% of KRAS mutations |
| G12V | Pancreatic, Colorectal | ~20% of KRAS mutations |
| G12C | Lung | ~45% of NSCLC KRAS mutations |
| G12R | Pancreatic | ~10-15% of PDAC mutations |
| G13D | Colorectal | ~14% of KRAS mutations |
| Q61H | Multiple | ~2% of KRAS mutations |
Classical Computational Approaches: Methodologies and Limitations
Molecular Dynamics and QM/MM Simulations
Classical molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) simulations have provided crucial insights into KRAS function and inhibition. Yan et al. utilized QM/MM simulations to elucidate the novel mechanism of GTP hydrolysis catalyzed by wild-type KRAS and the KRASG12R mutant [46]. Their methodology involved:
- System Preparation: Crystal structures of WT-KRAS-GTP and KRASG12R-GTP complexes (PDB IDs: 6XI7 and 6CU6) were obtained from the Protein Data Bank [46].
- Molecular Dynamics: Systems were solvated in TIP3P water boxes with NaCl concentration maintained at 0.15 M using AMBER20 package.
- QM/MM Calculations: Employed the ONIOM method with Gaussian 16 and AMBER20, with the QM region treated at the B3LYP/6-31G* level and MM region with the ff14SB forcefield.
- Pathway Analysis: Four distinct GTP hydrolysis mechanisms were computed and compared using potential energy surface scans [46].
This research revealed a novel GTP hydrolysis mechanism assisted by Mg²âº-coordinated water molecules, with energy barriers lower than previously reported pathways (14.8 kcal/mol for Model A and 18.5 kcal/mol for Model B) [46]. The G12R mutation was found to introduce significant steric hindrance at the hydrolysis site, explaining its impaired catalytic rate despite favorable energy barriers [46].
Structure-Based Virtual Screening
Structure-based virtual screening represents another workhorse classical approach. A 2022 study employed pharmacophore modeling, molecular docking, and MD simulations to identify KRAS G12D inhibitors [47]. The experimental protocol comprised:
- Pharmacophore Generation: A ligand-based common feature pharmacophore model was developed from known KRAS G12D inhibitors, featuring two hydrogen bond donors, one hydrogen bond acceptor, two aromatic rings, and one hydrophobic feature [47].
- Virtual Screening: Over 214,000 compounds from InterBioScreen and ZINC databases were screened against the pharmacophore model.
- Molecular Docking: Twenty-eight mapped compounds were docked against KRAS G12D, with four hits showing higher binding affinity than the reference inhibitor BI-2852.
- MD Validation: 100ns MD simulations confirmed stable binding and favorable free energy calculations for the top hits [47].
Performance Limitations of Classical Methods
While classical computational approaches have contributed significantly to KRAS drug discovery, they face fundamental limitations in scaling and accuracy. Classical force field-based docking struggles to capture KRAS's highly dynamic conformational landscape [48]. Molecular docking simulations are computationally expensive and frequently fail to scale across diverse chemical structures [49]. Deep learning models require large labeled datasets often scarce in drug discovery and struggle with high-dimensional molecular data, limiting generalization across different drug classes and target proteins [49].
Quantum Computing Approaches: Methodologies and Advantages
Quantum-Enhanced Generative Models
A landmark 2024 study published in Nature Biotechnology demonstrated a hybrid quantum-classical generative model for KRAS inhibitor design [44]. The workflow integrated three key components:
- Quantum Circuit Born Machine (QCBM): A 16-qubit processor generated prior distributions, leveraging quantum superposition and entanglement to explore chemical space.
- Long Short-Term Memory (LSTM) Network: A classical deep learning model for sequential data modeling.
- Chemistry42 Platform: Validation and refinement of generated structures [44].
The experimental protocol employed:
- Training Data Curation: Compiled 1.1 million data points from known KRAS inhibitors, VirtualFlow 2.0 screening of 100 million molecules, and STONED algorithm-generated compounds.
- Hybrid Model Training: QCBM generated samples from quantum hardware in each training epoch, trained with reward values calculated using Chemistry42.
- Compound Selection: Generated 1 million compounds screened for pharmacological viability and ranked by protein-ligand interaction scores [44].
This approach yielded two experimentally validated hit compounds (ISM061-018-2 and ISM061-022) demonstrating KRAS binding and functional inhibition in cellular assays [44]. The quantum-enhanced model showed a 21.5% improvement in passing synthesizability and stability filters compared to classical approaches, with success rates correlating approximately linearly with qubit count [44].
Quantum Kernel Methods for Drug-Target Interaction
The QKDTI (Quantum Kernel Drug-Target Interaction) framework represents another significant quantum advancement, employing Quantum Support Vector Regression (QSVR) with quantum feature mapping [49]. The methodology features:
- Quantum Feature Mapping: Transformation of classical biochemical features into quantum Hilbert spaces using parameterized RY and RZ-based quantum circuits.
- Nyström Approximation: Integration for efficient kernel approximation and reduced computational overhead.
- Parallel Quantum Kernel Computation: Batched pipeline for efficient processing of molecular samples [49].
Performance benchmarks on standard datasets demonstrated remarkable results, with accuracy rates of 94.21% on DAVIS, 99.99% on KIBA, and 89.26% on BindingDB, significantly outperforming classical models [49].
Quantum Lattice Boltzmann Method for Fluid Dynamics
While not directly applied to KRAS in the available literature, the Quantum Lattice Boltzmann Method (QLBM) represents an emerging quantum approach for simulating fluid dynamics at unprecedented scales [50]. Ansys and NVIDIA collaborated to execute a record-scale 39-qubit QLBM simulation using 183 nodes (1,464 GPUs) on the Gefion supercomputer, solving a problem with 68 billion degrees of freedom [50]. This demonstrates the massive scaling potential of quantum algorithms for complex physical simulations relevant to molecular dynamics.
Comparative Performance Analysis
Table 2: Quantum vs. Classical Computational Performance for KRAS Drug Discovery
| Performance Metric | Classical Approaches | Quantum-Hybrid Approaches | Experimental Context |
|---|---|---|---|
| Success Rate | Baseline | 21.5% improvement | Molecule generation passing synthesizability/stability filters [44] |
| Dataset Accuracy | <90% (DAVIS) | 94.21% (DAVIS) | Drug-target interaction prediction [49] |
| Binding Affinity Prediction | Variable across mutants | Pan-RAS activity demonstrated | ISM061-018-2 showed binding to multiple KRAS mutants [44] |
| Chemical Space Exploration | Limited by computational scaling | Efficient exploration of ~10â¶â° molecules | Quantum generative models [44] |
| Scalability | Linear with computational resources | Exponential in qubit count | 39-qubit simulation handling 68 billion degrees of freedom [50] |
| Experimental Validation | Multiple hits (e.g., [47]) | Two confirmed binders (ISM061-018-2, ISM061-022) | SPR and cell-based assays [44] |
Technical Implementation and Workflow Integration
Hybrid Quantum-HPC Infrastructure
Effective deployment of quantum workflows requires sophisticated integration with high-performance computing (HPC) infrastructure. The Quantum Framework (QFw) enables scalable hybrid quantum-HPC applications through [51]:
- Backend-Agnostic Execution: Unified interface for multiple quantum simulators (Qiskit Aer, NWQ-Sim, QTensor, TN-QVM) and hardware (IonQ).
- Workload-Optimized Routing: Circuit structure, entanglement, and depth determine optimal backend selection.
- Distributed Computation: Concurrent subproblem execution across HPC resources, demonstrated with NWQ-Sim's performance on large-scale entanglement and Hamiltonian simulations [51].
Quantum Algorithmic Advantages
The performance advantages of quantum approaches stem from fundamental physical principles:
- Quantum Superposition: Qubits represent multiple states simultaneously, enabling efficient exploration of chemical space.
- Quantum Entanglement: Correlations between qubits capture complex molecular interactions beyond classical capabilities.
- Quantum Interference: Amplifies correct solutions while canceling erroneous pathways [49] [45].
These properties allow quantum models to escape "barren plateaus" in optimization landscapes and represent complex probability distributions more efficiently than classical models [44].
Table 3: Essential Research Reagents and Computational Resources for KRAS Simulation
| Resource Name | Type | Function in Research | Example Use Case |
|---|---|---|---|
| NVIDIA CUDA-Q | Quantum Development Platform | Scalable GPU-accelerated quantum circuit simulations | QLBM implementation for fluid dynamics [50] |
| Chemistry42 | Software Platform | Validation of generated molecular structures | Pharmacological viability screening in hybrid workflow [44] |
| VirtualFlow 2.0 | Screening Platform | High-throughput virtual screening | Enamine REAL library screening for training data [44] |
| STONED Algorithm | Generative Algorithm | Generation of structurally similar compounds | Data augmentation for training set [44] |
| AMBER20 | Molecular Dynamics Package | Classical MD and QM/MM simulations | GTP hydrolysis mechanism study [46] |
| QCBM | Quantum Algorithm | Quantum generative modeling | Prior distribution generation in hybrid model [44] |
| Surface Plasmon Resonance | Experimental Validation | Binding affinity measurement | Confirmation of KRAS binding for generated compounds [44] |
| MaMTH-DS | Cell-Based Assay | Functional interaction monitoring | Dose-responsive inhibition testing across KRAS mutants [44] |
Visualization of Key Workflows and Signaling Pathways
Quantum-Classical Hybrid Workflow for KRAS Inhibitor Design
KRAS Signaling Pathway and Inhibition Mechanisms
Computational Scaling Paradigms: Classical vs. Quantum
The comparative analysis of quantum and classical computational workflows for KRAS inhibition reveals a rapidly evolving landscape where hybrid quantum-classical approaches are beginning to demonstrate measurable advantages in specific applications. Quantum-enhanced generative models have produced experimentally validated KRAS inhibitors that compare favorably with classically generated compounds, while quantum kernel methods show superior accuracy in drug-target interaction prediction [44] [49].
Nevertheless, classical approaches continue to provide crucial insights, as evidenced by QM/MM simulations elucidating fundamental KRAS biochemical mechanisms [46]. The optimal path forward appears to leverage the respective strengths of both paradigms: classical methods for well-characterized systems where force field accuracy is sufficient, and quantum approaches for exploring complex chemical spaces and modeling quantum mechanical effects in drug-target interactions.
As quantum hardware continues to advance and algorithmic innovations address current limitations in noise and qubit coherence, the scaling advantages predicted by quantum information theory may increasingly translate to practical drug discovery applications. For researchers targeting challenging systems like KRAS, maintaining expertise in both classical and quantum computational methodologies will be essential for leveraging the most appropriate tools for each stage of the drug discovery pipeline.
Quantum Computing for Protein Folding and Hydration Analysis
Classical computers face a fundamental scaling problem when simulating quantum mechanical systems in chemistry and biology. The resource requirements for exact simulations grow exponentially with the size of the molecular system, making problems like protein folding and hydration analysis computationally intractable for all but the smallest molecules [1]. Quantum computing offers a potential pathway to overcome this bottleneck by leveraging the inherent quantum properties of qubits—superposition and entanglement—to simulate nature with nature itself [52] [53].
This comparison guide examines the current landscape of quantum computing applications for protein folding and hydration analysis, focusing on direct performance comparisons between quantum and classical approaches. The field is rapidly evolving from theoretical promise to tangible demonstrations, with several recent breakthroughs indicating a trajectory toward practical quantum advantage in computational chemistry and drug discovery.
Performance Comparison: Quantum vs. Classical Approaches
Quantitative Performance Metrics
Table 1: Performance Comparison of Protein Folding Simulations
| Computing Approach | Maximum Problem Size Demonstrated | Algorithm/Method | Hardware Platform | Reported Accuracy/Performance |
|---|---|---|---|---|
| Quantum (Trapped-ion) | 12 amino acids [54] [55] | BF-DCQO [56] | IonQ Forte (36 qubits) [55] | Optimal solutions for all tested peptides [56] |
| Quantum (Superconducting) | 7-amino acid neuropeptide [53] | VQE with CVaR [53] | IBM Quantum Processor [53] | Reproducible results matching classical predictions [53] |
| Classical (AI-based) | Hundreds of amino acids [1] | AlphaFold2, RoseTTAFold | GPU Clusters | Near-experimental accuracy for many targets |
| Classical (Molecular Dynamics) | Dozens of amino acids [1] | Density Functional Theory | Supercomputers | Approximate solutions with accuracy trade-offs |
Table 2: Performance in Hydration and Binding Analysis
| Computing Approach | Application Focus | Method | Key Advantage |
|---|---|---|---|
| Quantum-Classical Hybrid | Protein hydration water placement [52] | Hybrid quantum-classical approach [52] | Precise water mapping in occluded protein pockets [52] |
| Quantum-Classical Hybrid | Ligand-protein binding studies [52] | Quantum-powered binding simulations [52] | Accurate modeling of water-mediated binding interactions [52] |
| Classical | Hydration analysis [1] | Molecular Dynamics | Well-established but computationally limited |
Analysis of Computational Scaling
The fundamental advantage of quantum computing lies in its scaling properties for specific problem classes. Protein folding represents an NP-hard combinatorial optimization problem whose complexity grows exponentially with chain length on classical computers [53]. Quantum approaches like the BF-DCQO algorithm demonstrate polynomial scaling for the same problem classes, potentially overcoming the exponential wall that limits classical methods [56].
For hydration analysis, classical methods struggle with the quantum mechanical nature of water molecules and their interactions with protein surfaces. The hybrid quantum-classical approach developed by Pasqal and Qubit Pharmaceuticals demonstrates more efficient mapping of water distributions within protein cavities, particularly in challenging occluded regions where classical sampling methods require prohibitive computational resources [52].
Experimental Protocols and Methodologies
Protein Folding on Trapped-Ion Quantum Computers
Experimental Protocol (IonQ & Kipu Quantum)
The record-breaking protein folding demonstration followed a structured workflow:
Problem Formulation: Protein sequences were mapped onto a tetrahedral lattice, with each amino acid's position encoded using two qubits, representing four possible directions for the chain to extend [54] [56].
Hamiltonian Construction: The energy function incorporated three key components:
- Geometric constraints preventing chain overlap
- Chirality terms ensuring proper stereochemistry
- Interaction energies based on known amino acid contact potentials [56]
Algorithm Implementation: The BF-DCQO (Bias-Field Digitized Counterdiabatic Quantum Optimization) algorithm was employed, which iteratively steers the quantum system toward lower energy states using dynamically updated bias fields [54] [55].
Hardware Execution: Problems were executed on IonQ's 36-qubit Forte quantum processor utilizing the inherent all-to-all connectivity of trapped-ion architecture [55] [56].
Post-Processing: Near-optimal solutions from quantum processing were refined using classical greedy local search algorithms to mitigate measurement errors [54].
Diagram 1: Quantum Protein Folding Workflow. This workflow illustrates the complete process from protein sequence to folded structure validation using quantum algorithms.
Hybrid Quantum-Classical Approach for Hydration Analysis
Experimental Protocol (Pasqal & Qubit Pharmaceuticals)
The methodology for protein hydration analysis involves tight integration between classical and quantum processing:
Classical Pre-Processing: Initial water density maps are generated using classical molecular dynamics simulations to identify probable hydration sites [52].
Quantum Refinement: Quantum algorithms precisely place water molecules within protein pockets, including regions that are challenging for classical sampling due to geometric constraints [52].
Binding Analysis: Water-mediated protein-ligand interactions are modeled using quantum principles to accurately simulate the binding thermodynamics under biologically relevant conditions [52].
The quantum hydration approach specifically leverages superposition to evaluate numerous water configurations simultaneously, providing more comprehensive sampling of the hydration landscape than classical Monte Carlo methods [52].
Quantum-Centric Supercomputing for Complex Chemical Systems
Experimental Protocol (Caltech & IBM)
The hybrid approach for complex chemical systems demonstrates how quantum and classical resources can be strategically combined:
Quantum Pre-Screening: An IBM quantum device with Heron processor (utilizing up to 77 qubits) identifies the most important components in the Hamiltonian matrix of an iron-sulfur cluster [27].
Classical Exact Solution: The reduced Hamiltonian is transferred to the Fugaku supercomputer for exact diagonalization and wave function calculation [27].
Validation: Results for the [4Fe-4S] molecular cluster are compared against classical heuristic methods, demonstrating the quantum-guided approach provides more rigorous selection of relevant matrix elements than classical approximation methods [27].
Diagram 2: Hybrid Quantum-Classical Computational Workflow. This diagram shows the integration of quantum screening with classical processing for solving complex chemical systems.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Essential Research Tools for Quantum Computational Chemistry
| Tool/Platform | Type | Primary Function | Key Features |
|---|---|---|---|
| IonQ Forte [54] [55] | Hardware | Trapped-ion quantum computer | All-to-all qubit connectivity, 36+ qubits |
| BF-DCQO [54] [56] | Algorithm | Quantum optimization | Non-variational, counterdiabatic controls |
| VQE with CVaR [53] | Algorithm | Ground state energy estimation | Focuses on low-energy tail of distribution |
| Qoro Divi SDK [53] | Software | Quantum algorithm development | Automated parallelization, circuit packing |
| QC-AFQMC [5] | Algorithm | Force calculation for molecular dynamics | Accurate atomic-level force computation |
| Hybrid Quantum-Classical [52] | Framework | Hydration analysis | Combines classical MD with quantum placement |
| Furo[2,3-b]pyridine-6-carbonitrile | Furo[2,3-b]pyridine-6-carbonitrile|High-Quality RUO | Bench Chemicals | |
| Bach-EI hydroboration reagent 2.0M | Bach-EI hydroboration reagent 2.0M, CAS:180840-34-0, MF:C11H17BN, MW:174.07 g/mol | Chemical Reagent | Bench Chemicals |
Discussion and Future Outlook
Current Limitations and Hardware Requirements
Despite promising demonstrations, current quantum approaches face significant scalability challenges. Modeling biologically relevant proteins typically requires thousands to millions of qubits [1]. For instance, Google estimated that approximately 2.7 million physical qubits would be needed to model the iron-molybdenum cofactor (FeMoco) involved in nitrogen fixation [1]. Current hardware with ~100 qubits remains insufficient for direct industrial application without sophisticated error mitigation and hybrid approaches.
Quantum hardware is also fragile, with qubits susceptible to decoherence and noise that limit circuit depth and fidelity [1]. Algorithm development must account for these hardware constraints through techniques like circuit pruning and error-robust ansatz design [54].
Path to Quantum Advantage
The path to unambiguous quantum advantage in protein folding and hydration analysis requires simultaneous progress across multiple fronts:
Hardware Scaling: IonQ's roadmap targeting 2 million qubits by 2030 represents the aggressive scaling needed for practical applications [5].
Algorithm Refinement: Non-variational algorithms like BF-DCQO that avoid "barren plateaus" represent promising directions for near-term applications [54] [56].
Hybrid Frameworks: Quantum-centric supercomputing, as demonstrated by Caltech and IBM, provides an immediate pathway to extract value from current quantum resources while hardware continues to develop [27].
As quantum hardware matures and algorithmic efficiency improves, quantum computing is positioned to fundamentally reshape computational chemistry and drug discovery, potentially reducing discovery timelines from years to months while enabling the precise molecular design that remains elusive with classical methods alone [52].
Overcoming Quantum Noise: Error Correction and Algorithmic Optimization on Today's Hardware
For computational chemistry, the path to simulating complex molecular systems with high fidelity is fraught with fundamental challenges on classical hardware. As the year-to-year gains in classical computer performance taper off, quantum computing offers a potential route to greater computational performance for problems in electronic structure, chemical quantum dynamics, and materials design [57]. Molecules are inherently quantum systems, and quantum computers can, in theory, simulate any part of a quantum system's behavior without the approximations required by classical methods like density functional theory [1]. However, the realization of this potential is critically dependent on overcoming a fundamental constraint: the fragility of quantum bits (qubits) to errors.
Current quantum devices fall within the noisy intermediate-scale quantum (NISQ) era, where qubit fidelity is limited by various error sources. For chemistry applications, which may require millions of qubits to model complex systems like cytochrome P450 enzymes or iron-molybdenum cofactor (FeMoco), these errors present a significant barrier to practical utility [1]. This guide objectively compares two pivotal strategies for mitigating different classes of quantum errors: Dynamical Decoupling for suppressing idling errors and Measurement Error Mitigation for addressing readout inaccuracies. We evaluate their performance across different hardware platforms, provide detailed experimental methodologies, and contextualize their importance for scaling quantum computational chemistry beyond classical limitations.
Dynamical Decoupling: Silencing Idling Qubits
Theoretical Foundation and Comparison of DD Sequences
Dynamical Decoupling (DD) is perhaps the simplest and least resource-intensive error suppression strategy for improving quantum computer performance [58]. It mitigates idling errors—errors that occur when a qubit is idle and not actively undergoing operations—by applying a specific sequence of single-qubit pulses that effectively average out the qubit's interaction with its noisy environment [59].
The fundamental principle originates from nuclear magnetic resonance (NMR) spectroscopy: by frequently flipping the qubit with control pulses, the effect of low-frequency environmental noise can be cancelled out. For a qubit exposed to a slow noise field, a simple spin echo sequence (a single π-pulse between two idle periods) can reverse the accumulation of phase error. Advanced DD sequences extend this concept with more complex pulse patterns to cancel higher-order errors [58].
Table 1: Comparison of Dynamical Decoupling Sequences and Performance
| DD Sequence | Key Characteristics | Pulse Order | Reported Performance Improvement |
|---|---|---|---|
| Basic Sequences (CPMG, XY4) | Traditional, simple structure [58] | Lower-order error cancellation | Can nearly match high-order sequences with optimized pulse timing [58] |
| Uhrig DD (UDD) | Asymmetrically spaced pulses [58] | High-order | Consistently high performance across devices [58] |
| Quadratic DD (QDD) | Built-in robustness to pulse imperfections [58] | High-order | Generally outperforms basic sequences [58] |
| Universally Robust (UR) | Designed for universal noise suppression [58] | High-order | Among the best performing sequences [58] |
| Adaptive DD (ADAPT) | Software framework; applies DD selectively based on program [59] | Program-dependent | 1.86x average (up to 5.73x) fidelity improvement vs. no DD; 1.2x vs. blanket DD [59] |
Experimental Protocols and Performance Data
Implementing DD requires embedding sequences of control pulses (typically π-pulses) during qubit idling periods. The performance varies significantly with the choice of sequence, pulse spacing, and hardware characteristics.
Key Experimental Protocol [58]:
- Sequence Selection: Choose a DD sequence from the available options (e.g., CPMG, XY4, UDD, QDD, UR).
- Open-Pulse Control: Utilize open-pulse functionality to precisely control the timing and application of pulses. The circuit-level implementation can be suboptimal and must be accounted for.
- Pulse Interval Optimization: The time between pulses (pulse interval) is a critical parameter. Surprisingly, the optimal interval is often substantially larger than the minimum possible interval on a given device.
- State Preservation Assessment: A common metric is the sequence's ability to preserve an arbitrary single-qubit state over time, measured by state fidelity.
The ADAPT framework provides an intelligent approach to DD implementation [59]. Its methodology is:
- Efficacy Estimation: Use a structurally similar "Decoy Circuit" with a known solution to estimate the efficacy of DD for each qubit.
- Subset Selection: Judiciously apply DD only to the subset of qubits that benefit most, rather than naively enabling it for all idle qubits.
- Localized Search: Employ a linear-complexity algorithm to avoid an exponential search over all possible qubit combinations.
Performance surveys across superconducting-qubit IBMQ devices show that high-order sequences like UR and QDD generally outperform basic ones, though optimizing the pulse interval for basic sequences can make their performance nearly match the high-order sequences [58]. ADAPT demonstrates that a targeted approach is superior, improving application fidelity by an average of 1.86x compared to no DD and by 1.2x compared to applying DD to all qubits [59].
Measurement Error Mitigation: Achieving Readout Fidelity
Beyond Simple Readout: The Challenge of Mid-Circuit Measurements
High-fidelity mid-circuit measurements (MCMs) are a critical component for useful quantum computing. They enable fault-tolerant quantum error correction, dynamic circuits, and are essential for solving classically intractable problems in chemistry and other fields [60]. Unlike terminal measurements that end a circuit, MCMs read out specific qubits without destroying them or disrupting their neighbors, allowing for subsequent conditional operations.
However, MCMs introduce their own error sources, particularly measurement-induced crosstalk, where the act of measuring one qubit introduces errors in unmeasured, neighboring qubits [60]. Few methods existed to comprehensively assess MCM performance until the recent development of the Quantum Instrument Randomized Benchmarking (QIRB) protocol. This protocol is the first fully scalable method for quantifying the combined rate of all errors in MCM operations [60].
Error Mitigation Protocols and Chemistry-Specific Techniques
QIRB Protocol [60]:
- Circuit Construction: Generate random Clifford circuits that integrate MCMs naturally within them. The processor's state is ideally stabilized by every MCM.
- Pauli Tracking: Use Pauli-tracking techniques to divide the possible output strings into "success" and "fail" subsets of equal size.
- Error Metric Calculation: For a circuit C, the metric is F = (Nsuccess - Nfail)/N, where N is the number of repetitions.
- Error Rate Extraction: Average F over many depth-d circuits and fit the decay to an exponential to extract the QIRB error rate (r_Ω), which is directly comparable to gate error rates from standard RB.
This protocol has been used to detect and eliminate previously undetected measurement-induced crosstalk in a 20-qubit trapped-ion quantum computer and to quantify how much of that error is eliminated by dynamical decoupling on a 27-qubit IBM processor [60].
For quantum chemistry algorithms like the Variational Quantum Eigensolver (VQE), specialized error mitigation techniques have been developed. Reference-state Error Mitigation (REM) is a cost-effective, chemistry-inspired method, but its effectiveness is limited for strongly correlated systems [61]. To address this, Multireference-state Error Mitigation (MREM) has been introduced. MREM systematically captures hardware noise in strongly correlated ground states by utilizing compact wavefunctions composed of a few dominant Slater determinants, engineered to have substantial overlap with the target ground state [61]. This approach has demonstrated significant improvements in computational accuracy for molecular systems like H2O, N2, and F2 compared to the original REM method [61].
Comparative Analysis in Chemical Computation
Performance Benchmarks and Trade-offs
Table 2: Error Mitigation Performance in Chemical Computations
| Method / Platform | Targeted Error | Chemistry Application Demonstrated | Reported Impact / Accuracy |
|---|---|---|---|
| ADAPT DD (IBMQ) [59] | Idling errors | General application-level fidelity | 1.86x avg (5.73x max) fidelity improvement over no DD |
| High-Order DD (UR, QDD) [58] | General decoherence | Arbitrary state preservation | Consistently high performance across superconducting devices |
| DD on MCMs (IBMQ) [60] | Measurement-induced crosstalk | General dynamic circuits | Quantifiably eliminated a portion of MCM-induced crosstalk error |
| MREM (Simulation) [61] | General hardware noise | Strongly correlated molecules (H2O, N2, F2) | Significant accuracy improvement over REM |
| IonQ QC-AFQMC [5] | Algorithmic precision | Atomic-level force calculations for carbon capture | More accurate than classical force methods |
The data reveals several trade-offs. DD is low-cost and widely applicable but requires careful sequence and parameter tuning [59] [58]. Chemistry-specific mitigation like MREM promises greater accuracy for its target problems but may be less general [61]. Furthermore, the choice of quantum algorithm is crucial. For instance, IonQ's implementation of the quantum-classical auxiliary-field quantum Monte Carlo (QC-AFQMC) algorithm demonstrated accurate computation of atomic-level forces, which is critical for modeling chemical reactivity and materials for carbon capture [5]. This goes beyond isolated energy calculations, enabling the tracing of reaction pathways.
The Path to Quantum Advantage in Chemistry
The integration of robust error mitigation is a prerequisite for achieving quantum advantage in chemistry. Useful industrial applications, such as modeling cytochrome P450 enzymes or designing novel catalysts, are estimated to require millions of physical qubits [1]. While current demonstrations, such as a 77-qubit simulation of an iron-sulfur cluster on an IBM Heron processor paired with a classical supercomputer, are groundbreaking, they have not yet definitively surpassed the best classical algorithms [27]. They do, however, provide a clear path forward. In this hybrid approach, the quantum computer identifies the most important components of a large Hamiltonian matrix, which is then solved exactly by a classical supercomputer, replacing classical heuristics with a more rigorous quantum-based selection [27].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Tools for Quantum Error Mitigation
| Tool / Resource | Function in Research | Relevance to Chemistry |
|---|---|---|
| Open-Pulse Control [58] | Enables precise timing and implementation of custom DD sequences. | Allows for fine-tuned protection of qubits during idle periods in complex molecular simulations. |
| Decoy Circuits [59] | Structurally similar circuits with known solutions used to test and optimize error mitigation strategies. | Provides a method to validate the setup for a specific chemistry problem before running the actual experiment. |
| QIRB Protocol [60] | A scalable benchmarking protocol to quantify errors introduced by mid-circuit measurements. | Critical for assessing the feasibility of quantum error correction in long, complex chemistry algorithms. |
| Givens Rotation Circuits [61] | Efficiently constructs quantum circuits to generate multi-reference states for error mitigation. | Key for implementing MREM to study strongly correlated electronic structures in molecules. |
| Quantum-Centric Supercomputing [27] | Hybrid architecture combining quantum processors with classical supercomputers. | Enables the decomposition of large chemical problems (e.g., [4Fe-4S] clusters) into tractable quantum and classical sub-tasks. |
| 3-Hydroxy-3',4'-dimethoxyflavone | 3-Hydroxy-3',4'-dimethoxyflavone|High-Purity Research Compound | |
| 3-Acetamido-6-nitrochromen-2-one | 3-Acetamido-6-nitrochromen-2-one, CAS:787-63-3, MF:C11H8N2O5, MW:248.19 g/mol | Chemical Reagent |
For researchers in chemistry and drug development, the accurate simulation of molecular systems remains a formidable challenge for classical computers. Problems involving strongly correlated electrons, such as modeling catalytic processes in nitrogenase or predicting the electronic properties of novel materials, often require approximations that limit accuracy [1]. Quantum computing promises to overcome these limitations by operating on the same quantum principles that govern molecular behavior, potentially enabling exact simulations of quantum systems currently beyond classical reach [1].
The fundamental obstacle on this path is quantum decoherence—the extreme fragility of quantum bits (qubits) that lose their quantum states due to minimal environmental interference. Logical qubits represent the solution: rather than relying on individual physical qubits, information is encoded across many physical qubits using quantum error correction codes, creating a fault-tolerant computational unit that preserves quantum information despite underlying hardware imperfections [62] [63]. This article provides a comparative analysis of leading approaches to building these logical qubits, examining experimental data and methodologies that demonstrate the rapid progress toward fault-tolerant quantum computing for chemical applications.
Quantum Error Correction: The Foundation of Logical Qubits
Core Principles and Challenges
Quantum error correction (QEC) creates stable logical qubits from multiple imperfect physical qubits by encoding quantum information redundantly. Unlike classical repetition codes, QEC must correct for errors without measuring the quantum information directly, using instead syndrome measurements that extract only error information [62]. The fundamental challenge lies in the quantum threshold theorem, which establishes that fault-tolerant quantum computation becomes possible when physical error rates fall below a specific threshold (approximately 0.01% to 1% depending on the code and noise model) [62] [63].
The 2025 Quantum Error Correction Report identifies real-time error correction as the "defining engineering hurdle" for the industry, shifting the bottleneck from qubit quality to the classical systems that must process millions of error signals per second and feed back corrections within microseconds [62]. This decoding challenge involves managing data rates "comparable to a single machine processing the streaming load of a global video platform every second" [62].
Leading Quantum Error Correction Codes
Table: Comparison of Leading Quantum Error Correction Approaches
| Code Type | Physical Requirements | Error Correction Overhead | Key Advantages | Leading Implementers |
|---|---|---|---|---|
| Surface Codes | Nearest-neighbor connectivity in 2D lattice | ~1000 physical qubits per logical qubit | High threshold (~1%), compatible with superconducting qubits | Google, IBM [62] |
| qLDPC Codes | Long-range connectivity between qubits | ~90% reduction in overhead compared to surface codes | High encoding rate, reduced physical qubit requirements | IBM [7] |
| Bosonic Codes | Harmonic oscillator modes with nonlinear element | Built-in protection against certain error types, hardware-efficient | Alice & Bob [64] | |
| Color Codes | 2D or 3D lattice with specific connectivity | Similar to surface codes | Transversal gates for more efficient computation | Academic researchers [65] |
Comparative Analysis of Hardware Platforms and Experimental Progress
Superconducting Qubit Approaches
IBM's Quantum Roadmap: IBM has demonstrated a complete hardware foundation for fault tolerance with its Quantum Loon processor, incorporating multiple routing layers for long-distance on-chip connections ("c-couplers") and qubit reset technologies [7]. Critically, IBM achieved real-time error decoding using qLDPC codes in less than 480 nanoseconds—a feat accomplished one year ahead of schedule that demonstrates the classical processing capability required for fault tolerance [7]. The company's Quantum Nighthawk processor, scheduled for deployment by end of 2025, features 120 qubits with 218 tunable couplers, enabling circuits with 30% more complexity than previous generations and supporting up to 5,000 two-qubit gates [7].
Google's Quantum AI team has pursued surface code implementations, with recent work focusing on dynamic circuit capabilities and the "Willow" chip that completed a benchmark calculation in approximately five minutes that would require a classical supercomputer 10 septillion years [66]. Google's demonstration of a "below-threshold memory system" marked a key milestone showing that textbook error correction designs could be reproduced in hardware at larger scales [62].
Neutral Atom Architectures
A Harvard-led collaboration demonstrated in 2025 a fault-tolerant system using 448 atomic qubits manipulated with an intricate sequence of techniques including physical entanglement, logical entanglement, and "quantum teleportation" to transfer quantum states between particles [63]. Their work, published in Nature, combined "all essential elements for a scalable, error-corrected quantum computation in an integrated architecture" [63]. The system implemented complex circuits with dozens of error correction layers, suppressing errors below the critical threshold where adding qubits further reduces errors rather than increasing them [63].
This neutral atom approach, developed in collaboration with QuEra Computing, uses rubidium atoms with lasers to reconfigure electrons into information-carrying qubits [63]. The team successfully created a system that is "conceptually scalable" toward larger quantum computers, with researchers noting that "by understanding the core mechanisms for enabling scalable, deep-circuit computation, you can essentially remove things that you don't need, reduce your overheads, and get to a practical regime much faster" [63].
Cat Qubit Innovation
Alice & Bob, in collaboration with NVIDIA, has pioneered cat qubits designed for inherent error resistance [64]. Their approach demonstrates potential to "reduce the hardware requirements for building a useful large-scale quantum computer by up to 200 times compared with competing approaches" [64]. The recent NVQLink architecture integrates quantum processing units with classical GPUs, delivering real-time orchestration for fault-tolerant applications through GPU compilation, live decoding, and dynamic calibration within unified quantum-classical workflows [64].
This cat qubit architecture creates qubits that are inherently protected against bit-flip errors, potentially reducing the overhead for quantum error correction significantly. Alice & Bob have "demonstrated experimental results surpassing those of technology giants such as Google or IBM" using this approach [64].
Experimental Data Comparison
Table: Experimental Performance Metrics for Logical Qubit Demonstrations
| Platform/Organization | Physical Qubit Count | Logical Qubit Demonstration | Key Error Metrics | Code Type |
|---|---|---|---|---|
| Harvard/QuEra [63] | 448 atomic qubits | Fault-tolerant system with dozens of error correction layers | Errors suppressed below critical threshold | Neutral atom codes |
| IBM [7] | 120 (Nighthawk) | Real-time decoding in <480 ns with qLDPC codes | qLDPC codes | |
| Google Quantum AI [62] [66] | 105 (Willow chip) | Below-threshold memory system, exponential error reduction | Surface codes | |
| Microsoft/Atom Computing [66] | 112 atoms | 28 logical qubits, 24 entangled logical qubits | 1,000-fold error reduction | Topological codes |
Experimental Protocols: Methodologies for Logical Qubit Creation
Workflow for Logical Qubit Encoding
The following diagram illustrates the generalized experimental workflow for creating and verifying logical qubits across different hardware platforms:
Diagram: Experimental Workflow for Logical Qubit Creation
Detailed Experimental Methodologies
Harvard/QuEra Neutral Atom Protocol: The Harvard team used arrays of rubidium-87 atoms trapped in optical tweezers, employing lasers to excite atoms into Rydberg states that facilitate controlled quantum interactions [63]. Their methodology involved:
- Initialization: Preparing all physical qubits in the ground state using optical pumping techniques
- Encoding: Applying precisely controlled laser pulses to create entanglement between atoms, encoding logical information across multiple physical qubits
- Syndrome Extraction: Implementing non-destructive measurements through collective fluorescence detection that reveals error patterns without collapsing logical quantum information
- Decoding: Using classical processing algorithms to interpret syndrome data and identify necessary corrections
- Correction: Applying quantum gates to physical qubits based on decoding results to eliminate errors in logical qubits
- Verification: Performing logical quantum tomography to characterize the performance and fidelity of the logical qubit operations [63]
IBM qLDPC Code Implementation: IBM's approach with quantum Low-Density Parity-Check codes focuses on reducing the resource overhead for error correction:
- Hardware Integration: Utilizing tunable couplers to create long-range connectivity between superconducting qubits essential for qLDPC implementation
- Real-time Decoding: Developing specialized classical hardware that processes error signals within the 480 nanosecond coherence window
- Parallel Qubit Operation: Implementing control systems that maintain synchronization across multiple logical qubit blocks
- Calibration: Continuous automated tuning of qubit parameters to maintain optimal performance across the processor [7]
The Scientist's Toolkit: Essential Research Reagents and Materials
Table: Key Experimental Components for Logical Qubit Research
| Component/Reagent | Function in Experiment | Example Implementation |
|---|---|---|
| Optical Tweezers | Precise positioning and manipulation of individual atoms | Harvard/QuEra neutral atom array positioning [63] |
| Superconducting Qubit Chips | Physical implementation of qubits using Josephson junctions | IBM Nighthawk processor with tunable couplers [7] |
| Rydberg Excitation Lasers | Creating highly excited atomic states for quantum gates | Neutral atom platforms using 420nm and 1013nm lasers [63] |
| Cryogenic Systems | Maintaining ultra-low temperatures (10-15 mK) for superconductivity | Dilution refrigerators for superconducting qubit platforms |
| Arbitrary Waveform Generators | Precisely controlling timing and shape of quantum pulses | Creating complex quantum gate operations |
| High-Speed Digital Processors | Real-time decoding of error syndromes | NVIDIA GPU integration for Alice & Bob's cat qubits [64] |
| Parametric Amplifiers | Quantum-limited amplification for qubit readout | Enhancing signal-to-noise for syndrome measurements |
| Optical Cavities | Enhancing light-matter interaction for qubit control | Trapped ion and neutral atom systems for state detection |
| 9-Ethylanthracene | 9-Ethylanthracene, CAS:605-83-4, MF:C16H14, MW:206.28 g/mol | Chemical Reagent |
Implications for Computational Chemistry and Drug Development
The progression toward fault-tolerant logical qubits holds particular significance for computational chemistry and pharmaceutical research. Current classical computational methods, including density functional theory (DFT) and coupled cluster theory, struggle with molecular systems containing strongly correlated electrons—precisely the systems where quantum computers promise the greatest advantage [1] [67].
Recent research provides a projected timeline for quantum advantage in computational chemistry, suggesting that while classical methods will likely remain dominant for large molecule calculations for the foreseeable future, quantum computers may offer advantages for "highly accurate calculations on smaller to medium-sized molecules, those with tens or hundreds of atoms, within the next decade" [67]. Specific calculations such as Full Configuration Interaction (FCI) and Coupled Cluster with perturbative triplets (CCSD(T)) "will be the first to be surpassed by quantum algorithms, potentially within the early 2030s" [67].
Notably, a 2025 Caltech-IBM collaboration demonstrated a hybrid quantum-classical approach studying an iron-sulfur cluster ([4Fe-4S]) important in nitrogen fixation, using up to 77 qubits to simplify quantum chemistry calculations before completing them on a classical supercomputer [27]. This "quantum-centric supercomputing" approach represents a practical intermediate step toward fully quantum solutions for chemical problems [27].
The following diagram illustrates the relationship between physical qubit quality, error correction overhead, and computational capability for chemistry applications:
Diagram: Path from Qubit Quality to Chemical Applications
For pharmaceutical researchers, the implications are profound. Quantum computers could eventually simulate complex biological systems like cytochrome P450 enzymes or model drug-target interactions with unprecedented accuracy [1]. Early demonstrations include quantum simulations of protein folding and molecular energy calculations that suggest a pathway toward these more complex applications [1] [66].
The development of logical qubits represents the critical path toward fault-tolerant quantum computers capable of solving chemically relevant problems. Current experimental demonstrations across superconducting, neutral atom, and specialized cat qubit platforms show rapid progress in error correction capabilities, with multiple groups having demonstrated key components of the fault-tolerance roadmap.
While significant challenges remain—particularly in scaling to large numbers of logical qubits and reducing the overhead of error correction—the progress documented throughout 2025 suggests that useful quantum computations for chemistry applications may be achievable within the next decade. For researchers in chemistry and pharmaceutical development, now is the time to develop quantum literacy and explore hybrid quantum-classical algorithms that can leverage these emerging capabilities as logical qubit technologies continue their rapid advancement from laboratory demonstrations to practical computational tools.
In the pursuit of quantum advantage for computational chemistry, researchers face a fundamental constraint: the exponential scaling of quantum mechanical equations with system size. Classical computational methods, particularly for strongly correlated electrons in systems crucial to drug development and materials science, often rely on approximations that limit their accuracy. Quantum computing promises to overcome these limitations by efficiently simulating quantum systems, but current hardware imposes severe restrictions on qubit counts, circuit depth, and coherence times. Within this challenging landscape, two methodological approaches have emerged as essential for making chemical simulations feasible on both current and near-term quantum devices: active space approximation and circuit transpilation. Active space approximation reduces the computational problem to a manageable subset of electrons and orbitals, while circuit transpilation translates abstract quantum algorithms into hardware-executable instructions. This guide provides an objective comparison of these approaches, their performance trade-offs, and implementation protocols to inform research strategies in computational chemistry and drug development.
Theoretical Framework and Key Concepts
Active Space Approximation in Quantum Chemistry
The active space approximation addresses the exponential complexity of quantum chemical simulations by strategically partitioning the electronic structure problem. In this paradigm, a subset of electrons and orbitals—the "active space"—is selected for high-level quantum treatment, while the remaining "inactive" electrons are handled with more efficient classical methods. Formally, this approach constructs a fragment Hamiltonian that focuses computational resources on the chemically relevant regions:
[ \hat{H}^{\text{frag}} = \sum{uv} V{uv}^{\text{emb}} \hat{a}u^\dagger \hat{a}v + \frac{1}{2} \sum{uvxy} g{uvxy} \hat{a}u^\dagger \hat{a}x^\dagger \hat{a}y \hat{a}v ]
where the embedding potential (V_{uv}^{\text{emb}}) captures interactions between active and inactive subsystems [68]. This framework enables researchers to apply expensive quantum algorithms to manageable active spaces while embedding them in a classically-treated molecular environment, dramatically reducing quantum resource requirements without sacrificing accuracy in chemically important regions.
Quantum Circuit Transpilation
Circuit transpilation addresses the implementation gap between abstract quantum algorithms and physical hardware constraints. The transpilation process decomposes high-level quantum operations into native gate sets specific to target hardware while optimizing circuit depth and qubit allocation to minimize errors. This process is particularly crucial because current quantum devices exhibit limited qubit connectivity, necessitating the insertion of SWAP gates to enable interactions between non-adjacent qubits—significantly increasing circuit complexity and error susceptibility [69]. Sophisticated transpilation algorithms employ techniques including gate cancellation, pulse shaping, and error-aware routing to balance circuit fidelity with execution efficiency, creating hardware-optimized implementations that maximize the likelihood of successful computation on noisy intermediate-scale quantum (NISQ) devices.
Quantum Algorithms for Chemistry
The table below summarizes key quantum algorithms for computational chemistry and their resource characteristics:
Table 1: Quantum Algorithms for Chemical Simulation
| Algorithm | Key Principle | Resource Requirements | Best-Suited Applications |
|---|---|---|---|
| VQE (Variational Quantum Eigensolver) | Hybrid quantum-classical optimization of parameterized circuits [70] | Lower circuit depth but high measurement overhead: (O(M^4/\epsilon^2)) to (O(M^6/\epsilon^2)) measurements for M basis functions [71] | Near-term applications; ground state energy calculations [1] |
| QPE (Quantum Phase Estimation) | Quantum Fourier transform to extract energy eigenvalues [70] | High logical qubit counts (693+ for H₂O) and gate complexity (~10⸠gates) [71] | Fault-tolerant era; high-precision energy calculations |
| Qubitization | Hamiltonian embedding into unitary operators [70] | Polynomial scaling improvements: from (O(M^{11})) to (O(M^5)) for Gaussian orbitals [70] | Efficient Hamiltonian simulation in fault-tolerant setting |
Methodological Approaches and Protocols
Active Space Selection and Optimization Protocols
The quantum information-assisted complete active space optimization (QICAS) protocol represents a significant advancement in systematic active space selection. This approach leverages quantum information measures, particularly orbital entanglement entropy, to identify optimal active spaces with minimal empirical input:
[ S(\rhoi) = -\rhoi \log(\rhoi), \quad \rhoi = \text{Tr}{\backslash{\phii}}[|\Psi0\rangle\langle\Psi0|] ]
where (S(\rhoi)) quantifies the entanglement between orbital (\phii) and the rest of the system [72]. The QICAS protocol minimizes the "out-of-CAS correlation"—the sum of orbital entropies over all non-active orbitals—yielding optimized active spaces that capture essential correlation effects. Implementation involves (1) computing an approximate ground state (|\Psi_0\rangle) using efficient classical methods like density matrix renormalization group (DMRG) with low bond dimension, (2) calculating single-orbital entropies for all orbitals, (3) selecting orbitals with highest entropy values for the active space, and (4) iteratively optimizing the orbital basis to minimize discarded correlation [72]. This method has demonstrated exceptional performance, producing active spaces that approach CASSCF accuracy with CASCI calculations for small correlated molecules and significantly accelerating convergence for challenging systems like the chromium dimer.
Circuit Transpilation and Optimization Workflows
The quantum circuit transpilation process transforms algorithm-level quantum circuits into hardware-executable instructions through a multi-stage optimization pipeline:
Figure 1: Quantum Circuit Transpilation Workflow
The transpilation process begins with gate decomposition, translating abstract quantum operations into a device's native gate set (e.g., single-qubit rotations and CNOT gates for superconducting qubits). Next, qubit mapping assigns logical qubits to physical qubits, minimizing SWAP gate insertions required to overcome limited qubit connectivity—a critical step as SWAP gates triple the basic two-qu gate count and significantly increase error rates [69]. Gate scheduling then optimizes operation timing to minimize circuit depth and decoherence effects. Advanced transpilers incorporate hardware-specific characteristics including gate fidelity, qubit connectivity graphs, and coherence times through hardware-aware compilation, employing techniques like dynamical decoupling and error-aware routing to further enhance circuit performance [69].
Hybrid Quantum-Classical Embedding Methods
For complex chemical systems exceeding near-term quantum capabilities, hybrid quantum-classical embedding methods provide a practical pathway. The periodic range-separated DFT embedding approach demonstrates this paradigm, combining classical computational chemistry software (CP2K) with quantum algorithms (implemented in Qiskit Nature) through message-passing interfaces [68]. The protocol involves: (1) performing a classical DFT calculation of the entire system, (2) identifying the fragment (active space) for quantum treatment, (3) constructing the embedded fragment Hamiltonian with an effective embedding potential, (4) solving the fragment Hamiltonian using VQE or QPE on quantum hardware, and (5) iterating if necessary to achieve self-consistency. This approach was successfully applied to study the neutral oxygen vacancy in magnesium oxide, achieving accurate prediction of the experimental photoluminescence emission peak despite some discrepancies in the main absorption band position [68].
Performance Comparison and Experimental Data
Resource Requirements for Chemical Simulations
The table below quantifies quantum resource requirements for representative chemical simulations, highlighting the substantial reductions enabled by the methods discussed:
Table 2: Quantum Resource Estimates for Chemical Simulations
| System | Method | Logical Qubits | Non-Clifford Gates | Key Experimental Findings |
|---|---|---|---|---|
| H₂O (6-31g basis) | QPE with double factorization [71] | 693 | 3.06×10⸠| Target error 0.0016 Ha; 10× error increase reduces gates to 3.06×10ⷠ|
| Li₂FeSiO₄ (periodic) | QPE with first quantization [71] | 3,331 | 1.42×10¹ⴠ| 156 electrons, 10ⵠplane waves; massive gate complexity |
| [4Fe-4S] cluster | Hybrid quantum-classical [27] | 77 | N/A | Quantum computer identified important Hamiltonian components; classical solver computed exact wavefunction |
| O vacancy in MgO | Periodic rsDFT embedding [68] | Reduced active space | N/A | Accurate photoluminescence prediction vs. experiment; competitive with advanced ab initio methods |
Performance Trade-offs and Limitations
Both active space approximation and circuit transpilation introduce characteristic trade-offs between accuracy and efficiency:
Active Space Limitations: The accuracy of active space methods depends critically on selecting appropriate orbitals and electrons. Oversimplified active spaces miss crucial correlation effects, while excessively large active spaces exceed quantum resources. For example, even the [4Fe-4S] cluster—a biologically essential cofactor—required sophisticated hybrid approaches rather than full quantum simulation [27]. Industrial applications targeting cytochrome P450 enzymes or nitrogenase FeMoco clusters may require ~100,000 physical qubits—far beyond current capabilities [1].
Transpilation Overheads: Circuit optimization introduces its own resource costs. Transpilation of moderate-sized algorithms can require hours of classical computation time [69]. The compiled circuits often exhibit substantial gate overheads, particularly through SWAP networks that enable limited connectivity qubit architectures. For example, the Jordan-Wigner transformation—a common fermion-to-qubit mapping—introduces non-local string operators that significantly increase circuit depth [70]. Error mitigation techniques like zero-noise extrapolation and probabilistic error cancellation provide some compensation but further increase measurement overheads [69].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Computational Tools for Quantum Computational Chemistry
| Tool/Resource | Type | Primary Function | Application Example |
|---|---|---|---|
| CP2K [68] | Software Package | Ab initio molecular dynamics | Periodic DFT calculations for embedding environments |
| Qiskit Nature [68] | Quantum Algorithm Library | Quantum circuit ansatzes for chemistry | VQE and QPE implementation for active space problems |
| PennyLane [71] | Quantum ML Library | Resource estimation and algorithm development | Estimating logical qubits and gate counts for molecules |
| Double Factorization [71] | Algorithmic Technique | Hamiltonian representation compression | Reducing QPE gate counts from O(M¹¹) to O(Mâµ) |
| QICAS [72] | Orbital Selection Protocol | Correlation-driven active space selection | Optimal orbital identification for chromium dimer |
| Jordan-Wigner Encoding [70] | Qubit Mapping | Fermion-to-qubit transformation | Representing electronic orbitals as qubit states |
| FSWAP Networks [70] | Circuit Optimization | Efficient fermionic SWAP operations | Reducing overhead from non-local Jordan-Wigner strings |
For researchers and drug development professionals selecting computational strategies, the choice between active space approximation and circuit optimization depends on the specific chemical problem and available resources. Active space methods particularly benefit systems with localized strong correlation—such as transition metal active sites in enzymes—where chemical intuition or entropy-based metrics can guide orbital selection. Circuit optimization becomes crucial when pushing the limits of quantum hardware for a fixed active space size, maximizing feasible circuit depth through hardware-aware compilation.
The most promising path forward leverages both approaches synergistically: using active space selection to minimize quantum resource demands, then applying advanced transpilation to implement the resulting quantum algorithms with maximum efficiency. As quantum hardware continues to evolve, these resource reduction strategies will remain essential for bridging the gap between chemical complexity and computational feasibility, potentially enabling quantum advantage for targeted chemical applications in the coming decade.
Benchmarking Quantum vs. Quantum-Inspired Classical Algorithms
For researchers in chemistry and drug development, the competition between quantum and classical computing paradigms is intensifying. While fault-tolerant quantum computers promise revolutionary long-term potential, quantum-inspired classical algorithms currently deliver practical, scalable solutions for real-world problems. This guide provides an objective comparison of their performance, methodologies, and optimal application domains based on current experimental data, framing the analysis within the broader thesis of computational scaling in chemistry research.
Performance Benchmarking at a Glance
The table below summarizes key performance indicators from recent studies and industry demonstrations, highlighting the contrasting maturity and application profiles of these technologies.
| Algorithm / System | Application Domain | Performance Outcome | Key Metrics | Source / Experiment |
|---|---|---|---|---|
| Variational Quantum Algorithms (VQA) [73] [74] | Time Series Prediction | Underperformed vs. simple classical models | Accuracy on chaotic systems; model complexity | Comprehensive benchmark on 27 tasks [73] |
| Hybrid Quantum-Classical (IBM/Caltech) [27] | [4Fe-4S] Molecular Cluster | Beyond previous quantum limits, not yet definitively superior | 77 qubits used; quantum-assisted matrix simplification | Quantum-centric supercomputing [27] |
| IonQ 36-Qubit Computer [66] | Medical Device Simulation | 12% performance improvement over classical HPC | Real-world application speed and accuracy | Industry milestone demonstration [66] |
| Quantum-Inspired Algorithms [1] [66] | Optimization, Clean Hydrogen Catalyst Discovery | Effective on classical HPC; cannot fully replicate quantum | Speed and accuracy on specific problem classes | Fujitsu & Toshiba implementations [1] |
| HAWI (Hybrid Algorithm) [75] | Learning-With-Errors (LWE) Problem | Validated on 5-qubit device; potential for advantage | Qubit count < m(m+1); success probability | 2D problem solved on NISQ device [75] |
Experimental Protocols and Workflows
Protocol: Quantum-Centric Supercomputing for Molecular Simulation
This hybrid methodology, used to study the [4Fe-4S] molecular cluster, demonstrates the integrated use of quantum and classical resources [27].
- Problem Formulation: The chemical system is defined by inputting atomic positions, electron count, and other parameters into a classical algorithm.
- Hamiltonian Generation: The classical algorithm constructs the full Hamiltonian matrix, which grows exponentially with system size.
- Quantum Pre-processing: The Hamiltonian is transferred to a quantum processor (e.g., IBM Heron). The quantum computer identifies and returns the most crucial components of the matrix, replacing classical heuristics with a more rigorous selection [27].
- Classical Post-processing: The reduced Hamiltonian, enriched by quantum computation, is fed into a high-performance classical supercomputer (e.g., RIKEN's Fugaku) to solve for the exact wave function and ground-state energy [27].
The following workflow diagram illustrates these steps:
Protocol: Quantum-Inspired Algorithm for Classical HPC
This protocol outlines the general approach for developing and running quantum-inspired algorithms on classical hardware [1] [66].
- Algorithmic Abstraction: Analyze a quantum algorithm (e.g., VQE or QAOA) to identify its core computational principles, such as its approach to exploring a solution space.
- Classical Translation: Map these quantum principles onto efficient classical data structures and linear algebra operations, such as tensor networks or Monte Carlo methods.
- HPC Implementation: Optimize and run the translated algorithm on classical high-performance computing infrastructure, including GPUs and supercomputers.
- Validation & Benchmarking: Compare the results and computational cost (time, energy) against both the native quantum algorithm and other state-of-the-art classical methods.
The process is summarized in the following diagram:
The Scientist's Toolkit: Research Reagent Solutions
This section details the essential hardware and software components required for experiments in this field.
| Tool Name | Type | Function in Research |
|---|---|---|
| IBM Heron Processor [27] | Hardware (Quantum) | 127-qubit superconducting quantum processor; used for hybrid algorithm steps like Hamiltonian matrix simplification. |
| RIKEN Fugaku [27] | Hardware (Classical) | Supercomputer; handles the most computationally intensive post-processing steps in hybrid workflows. |
| Variational Quantum Eigensolver (VQE) [1] | Algorithm (Quantum) | A leading hybrid algorithm for estimating molecular ground-state energy on near-term devices. |
| Quantum Approximate Optimization Algorithm (QAOA) [75] | Algorithm (Quantum) | Used for combinatorial optimization problems; can be adapted for chemistry applications. |
| Quantum-Inspired Optimization [1] [66] | Algorithm (Classical) | Algorithms derived from quantum techniques but run on classical HPC to solve problems in optimization and catalyst discovery. |
| Error Mitigation Techniques [76] | Software/Methodology | Methods like dynamical decoupling and measurement error mitigation are crucial for obtaining meaningful results from current noisy quantum hardware. |
Analysis of Computational Scaling in Chemistry
The long-term thesis for quantum computing in chemistry rests on its superior scaling for simulating quantum systems. Classical methods like Density Functional Theory (DFT) often rely on approximations that fail for complex molecules with strong electron correlation, such as catalytic sites in enzymes or excited states in photochemistry [77]. Quantum computers, by contrast, are native quantum systems that can, in principle, simulate these problems with more favorable scaling, potentially unlocking new discoveries in drug and materials design [1].
However, this theoretical advantage is currently balanced by immense practical challenges. Current quantum devices are limited by qubit count, high error rates, and short coherence times [1]. For example, simulating industrially relevant molecules like cytochrome P450 enzymes is estimated to require millions of physical qubits [1]. This has created a window of opportunity for quantum-inspired classical algorithms, which leverage insights from quantum information to create better classical solvers for challenging problems like electronic structure calculation and molecular dynamics [1] [66].
The trajectory suggests a transitional era defined by hybrid quantum-classical approaches [27] [77]. In these workflows, quantum processors act as specialized co-processors for specific, computationally demanding sub-tasks—such as determining the most important components of a molecular Hamiltonian—while classical computers manage the overall workflow and post-processing [27]. This co-design is seen as the most viable path to achieving "quantum utility," where quantum computers deliver reliable results for scientifically meaningful chemistry problems ahead of full fault-tolerance [77].
Proof of Concept: Validating Unconditional Quantum Speedup in Chemistry-Relevant Problems
For researchers in chemistry and drug development, the simulation of molecular systems is a foundational yet formidable challenge. Classical computers struggle with the exact modeling of quantum mechanical phenomena, such as electron correlations in complex molecules, forcing reliance on approximations that limit accuracy and predictive power [1]. The core of this limitation is a scaling problem: the computational resources required for exact classical simulation grow exponentially with the size of the quantum system. Quantum computing, architected on the principles of quantum mechanics, promises to overcome this barrier by mimicking nature with native physics. This guide examines a pivotal milestone: the recent experimental demonstration of unconditional exponential quantum speedup. This achievement signals a potential paradigm shift, suggesting that for a specific, growing class of problems, quantum processors are embarking on a scaling trajectory that classical computers cannot follow [76].
Defining the Quantum Advantage
In the quest for practical quantum computing, "quantum advantage" is the critical benchmark. It is achieved when a quantum computer solves a problem faster or more accurately than any possible classical computer. A key distinction lies in the type of speedup:
- Polynomial Speedup: The quantum computer's advantage grows as a polynomial function of the problem size. This is valuable but offers a less dramatic performance separation.
- Exponential Speedup: The quantum computer's advantage grows as an exponential function of the problem size. For each additional variable (e.g., a qubit), the performance gap roughly doubles. This is the "most dramatic" type of speedup and leads to problems that become completely intractable for classical machines as they scale [76].
Furthermore, speedup can be conditional or unconditional:
- Conditional Speedup: Relies on unproven computational complexity assumptions, such as the belief that no better classical algorithm exists.
- Unconditional Speedup: Holds without any such assumptions, representing a fundamental and incontrovertible advantage [76].
The recent demonstration of unconditional exponential speedup marks a transition from theoretical promise to a tangible, scaling reality for quantum computing.
Experimental Protocols: Methodologies for a Quantum Speedup
The following experiments represent cutting-edge methodologies designed to prove quantum computational advantage.
The USC-IBM Experiment on Simon's Problem
This study was designed to demonstrate an unconditional exponential speedup by solving a variation of Simon's problem, a precursor to Shor's factoring algorithm [76].
Objective: To find a hidden repeating pattern in a black-box function. A quantum player can identify the secret pattern exponentially faster than any classical strategy [76].
- Quantum Hardware: Two 127-qubit IBM Quantum Eagle processors.
- Algorithm: A modified quantum algorithm for an Abelian Hidden Subgroup Problem.
- Key Error Mitigation Protocols:
- Circuit Compression & Input Restriction: Reduced the number of quantum logic gates, minimizing error accumulation [76].
- Dynamical Decoupling: Applied specific pulse sequences to shield qubits from environmental noise. This was cited as the most crucial factor in achieving the speedup [76].
- Measurement Error Mitigation: Corrected for imperfections in reading the final state of the qubits [76].
Google's Quantum Echoes Algorithm
This experiment aimed to demonstrate a beyond-classical capability for a complex physics simulation with links to real-world scientific tools like NMR spectroscopy [78].
Objective: To measure a subtle quantum interference effect known as the second-order out-of-time-order correlator (OTOC²) and use it for Hamiltonian learning [78].
- Quantum Hardware: A 65-qubit superconducting processor (Willow chip).
- Algorithm: The "Quantum Echoes" algorithm, which uses a time-reversal (echo) protocol.
- Key Experimental Workflow:
- Forward Evolution: The quantum system evolves for a set time.
- Butterfly Perturbation: A small, precisely applied disturbance.
- Backward Evolution: The system's evolution is reversed in time.
- Interference Measurement: The resulting quantum interference is measured, revealing information about chaos and scrambling in the system [78].
- Application to NMR: The same technique was shown to theoretically extend the effective range of nuclear magnetic resonance (NMR) spectroscopy, creating a "longer molecular ruler" for determining molecular structures [78].
IonQ & Ansys: A Practical Application Benchmark
This experiment focused on demonstrating a quantum utility advantage for a specific engineering problem [66] [79].
Objective: To speed up the simulation of fluid interactions in a medical device component [79].
- Quantum Hardware: IonQ's 36-qubit quantum computer.
- Methodology: A hybrid quantum-classical algorithm was used to model the fluid dynamics.
- Benchmarking: The results were compared directly against simulations run on classical high-performance computing (HPC) clusters [79].
The logical pathway and key decision points for achieving quantum speedup are summarized in the diagram below.
Comparative Performance Data
The quantitative results from these advanced experiments provide compelling evidence of quantum computing's growing capabilities. The following tables summarize the key performance metrics and outcomes.
Table 1: Experimental Protocols and Key Outcomes
| Experiment | Primary Objective | Quantum Hardware | Key Algorithm/Protocol |
|---|---|---|---|
| USC-IBM (Simon's Problem) [76] | Demonstrate unconditional exponential scaling advantage | 127-qubit IBM Eagle Processor | Modified Abelian Hidden Subgroup Algorithm |
| Google (Quantum Echoes) [78] | Measure OTOC² and demonstrate verifiable speedup | 65-qubit Willow Processor | Quantum Echoes (Time-Reversal) Algorithm |
| IonQ & Ansys [79] | Outperform classical HPC in a fluid dynamics simulation | IonQ 36-qubit System | Hybrid Quantum-Classical Algorithm |
Table 2: Quantitative Results and Classical Comparison
| Experiment | Reported Quantum Performance | Classical Benchmark & Performance | Speedup / Advantage |
|---|---|---|---|
| USC-IBM (Simon's Problem) [76] | Successful execution with unconditional scaling | Any classical algorithm | Unconditional Exponential Scaling Advantage |
| Google (Quantum Echoes) [78] | 2.1 hours for 65-qubit OTOC² calculation | Frontier supercomputer: estimated 3.2 years | ~13,000x speedup |
| IonQ & Ansys [79] | Accurate simulation completed | Classical HPC simulation | 12% performance improvement |
The Scientist's Toolkit: Essential Research Reagents
For researchers seeking to understand or replicate work at the quantum-classical frontier, the following "research reagents"—the core components and techniques—are essential.
Table 3: Key Reagents for Quantum Speedup Experiments
| Research Reagent | Function & Role | Example in Context |
|---|---|---|
| Noise Intermediate-Scale Quantum (NISQ) Processors | The physical quantum hardware that executes algorithms; characterized by high but improving qubit counts and error rates. | IBM's 127-qubit Eagle [76], Google's 65-qubit Willow [78]. |
| Dynamical Decoupling | A pulse sequence technique that protects qubits from decoherence by decoupling them from a noisy environment. | Critical for achieving speedup in the USC experiment [76]. |
| Measurement Error Mitigation | A classical post-processing technique that corrects for readout errors at the end of a quantum computation. | Used in both USC and Google experiments to improve result fidelity [76]. |
| Transpilation | The process of compiling a high-level quantum circuit into the specific, native gate set of a target quantum processor. | Used in the USC experiment to compress circuits and reduce gate count [76]. |
| Time-Reversal (Echo) Protocols | Core component of algorithms that study quantum chaos and scrambling by running evolution forward and backward. | The foundation of Google's Quantum Echoes algorithm for measuring OTOC² [78]. |
| High-Coherence Qubits | Qubits with long coherence times (Tâ‚‚), enabling more complex computations before information is lost. | Aalto University achieved a record ~1 ms coherence, reducing error correction burden [80]. |
The workflow for Google's Quantum Echoes algorithm, which connects a core quantum protocol to a practical chemical application, is illustrated below.
Implications for Chemistry and Drug Development
The demonstrated exponential quantum speedup, while currently applied to abstract problems, charts a clear course toward transformative applications in chemistry and pharmacology. Quantum computers are inherently suited to simulate molecular and electronic quantum states without the approximations required by classical methods like density functional theory (DFT) [1]. This capability could precisely model phenomena such as:
- Strongly correlated electron systems, which are crucial in catalysis and materials science [1] [81].
- Complete reaction pathways and transition states, accelerating the design of novel catalysts.
- Direct simulation of complex biomolecules, such as cytochrome P450 enzymes and the iron-molybdenum cofactor (FeMoco) involved in nitrogen fixation—tasks that are currently beyond the reach of exact classical simulation [1] [66].
While current AI methods have made impressive strides in approximating quantum chemical properties for large, weakly correlated systems [81], the emergence of unconditional exponential quantum scaling addresses a fundamentally different problem class. For the complex, strongly correlated quantum systems at the heart of many unsolved problems in drug discovery and materials design, quantum computing offers a scalable path to solutions that may forever remain out of reach for purely classical machines. The future likely lies in hybrid quantum-classical AI, where each technology handles the tasks to which it is best suited [81].
A central thesis in modern computational science is that quantum computers offer a fundamental advantage over classical systems for specific, critically important problems. This advantage is not merely a linear speedup but an exponential reduction in computational complexity, transforming problems from intractable to manageable. This case study examines this thesis through two distinct lenses: the applied challenge of simulating complex chemical systems, specifically iron-sulfur clusters, and the foundational computational problem of solving the Abelian Hidden Subgroup Problem (HSP). The former represents a direct application with immense implications for chemistry and drug discovery, while the latter provides the mathematical underpinning for the quantum algorithms that enable such applications.
Classical computing methods, including density functional theory, struggle with the accurate simulation of quantum systems because the resources required grow exponentially with the size of the system [1]. This is particularly true for molecules with strong electron correlations, such as the iron-sulfur clusters prevalent in metabolic enzymes [1] [82]. Similarly, the best-known classical algorithms for problems equivalent to the Abelian HSP require a number of steps that grows exponentially with the problem size, while quantum algorithms require only polynomially more steps [83]. This case study will objectively compare the performance of quantum, classical, and hybrid approaches against experimental data, detailing the protocols that define the state of the art.
Quantum-Centric Supercomputing for Iron-Sulfur Clusters
Experimental Protocol: Hybrid Quantum-Classical Workflow
A 2025 study by Caltech, IBM, and RIKEN established a new benchmark for simulating chemical systems by using a quantum-centric supercomputing approach to study the [4Fe-4S] molecular cluster, a critical component in enzymes like nitrogenase [27]. The detailed experimental methodology is as follows:
- Step 1 — Problem Formulation: The electronic structure of the [4Fe-4S] cluster is encoded into a mathematical representation known as the Hamiltonian matrix. On a classical computer, this matrix becomes exponentially large and intractable for exact solution.
- Step 2 — Quantum-Driven Truncation: An IBM quantum device, powered by a Heron processor, is employed to identify the most relevant components of the Hamiltonian. The quantum computer performs a rigorous selection, replacing the classical heuristics typically used to approximate and prune the matrix [27].
- Step 3 — Classical Exact Solution: The truncated, more manageable Hamiltonian is then passed to the RIKEN Fugaku supercomputer, one of the world's most powerful classical computers, which calculates the exact ground-state wave function and energy [27].
- Step 4 — Validation and Analysis: The resulting wave function is analyzed to determine properties such as reactivity and stability, which are validated against known chemical behavior of iron-sulfur clusters.
This protocol used up to 77 physical qubits on the quantum processor, significantly more than most prior quantum chemistry experiments [27].
Performance & Scaling Comparison
The table below summarizes the performance data and scaling characteristics of different computational approaches for simulating the [4Fe-4S] cluster.
Table 1: Performance comparison for [4Fe-4S] cluster simulation
| Computational Approach | Key Method | Qubit Count | Classical Processing | Performance Outcome |
|---|---|---|---|---|
| Quantum-Centric Supercomputing (Caltech/IBM, 2025) | Quantum processor truncates Hamiltonian; supercomputer finds exact solution [27]. | 77 | RIKEN Fugaku Supercomputer | Produced chemically useful results beyond the reach of standard classical algorithms [27]. |
| Pure Classical Heuristics | Approximates Hamiltonian using classical algorithms [27]. | N/A | High-Performance Computing | Struggles with correct wave function; accuracy is unreliable [27]. |
| Theoretical Fault-Tolerant Quantum Computing | Full quantum simulation with error-corrected qubits. | ~100,000 (estimated) | Minimal | Required for full simulation of complexes like Cytochrome P450 [1]. |
The Researcher's Toolkit: Iron-Sulfur Cluster Simulation
Table 2: Essential research reagents and tools for quantum simulation of iron-sulfur clusters
| Research Tool | Function in the Experiment |
|---|---|
| Heron Quantum Processor (IBM) | Executed quantum algorithms to identify and truncate the most relevant parts of the large Hamiltonian matrix [27]. |
| Fugaku Supercomputer (RIKEN) | Solved the complex quantum chemistry problem exactly using the truncated Hamiltonian provided by the quantum processor [27]. |
| Hamiltonian Matrix | A mathematical representation that encapsulates all the energy levels and interactions of the electrons in the system [27]. |
| [4Fe-4S] Cluster Model | A model of the iron-sulfur protein cofactor, an essential benchmark for its complexity and biological importance [27]. |
Workflow Visualization
The Abelian Hidden Subgroup Problem
Problem Statement and Quantum Protocol
The Hidden Subgroup Problem (HSP) is a foundational framework in quantum computing. Given a group ( G ) and a function ( f ) that is constant and distinct on the cosets of an unknown subgroup ( H ), the task is to find ( H ) [84] [83]. For finite Abelian (commutative) groups, quantum computers provide an efficient solution.
The standard quantum algorithm for the Abelian HSP, which generalizes Shor's and Simon's algorithms, follows this protocol [84] [83]:
- Step 1 — State Preparation: Initialize two quantum registers. Apply a Hadamard transform to the first register to create a uniform superposition over all group elements: [ \frac{1}{\sqrt{|G|}}\sum_{g\in G}|g\rangle|0\rangle ]
- Step 2 — Oracle Query: Apply the function ( f ) via a quantum oracle, which entangles the two registers: [ \frac{1}{\sqrt{|G|}}\sum_{g\in G}|g\rangle|f(g)\rangle ]
- Step 3 — Measurement: Measure the second register. This collapses the first register into a superposition over a random coset of ( H ), yielding the state [ \frac{1}{\sqrt{|H|}}\sum_{h\in H}|s + h\rangle ] for some random ( s \in G ).
- Step 4 — Quantum Fourier Transform (QFT): Apply the QFT over ( G ) to the first register. This transforms the state from the coset superposition into a state that reveals information about ( H^\perp ), the orthogonal subgroup of ( H ).
- Step 5 — Measurement and Repetition: Measure the first register to sample a random element from ( H^\perp ). By repeating this process ( O(\log |G|) ) times, the subgroup ( H ) can be determined with high probability [84] [83].
Recent advancements, such as the "initialization-free" algorithm by Kwon and Kim (2025), build on this standard method by removing the need to re-initialize the auxiliary register, thereby improving efficiency [85].
Instances and Performance Scaling
The following table compares key instances of the Abelian HSP and their quantum solutions.
Table 3: Quantum algorithms for Abelian Hidden Subgroup Problems
| Problem Instance | Group ( G ) | Hidden Subgroup ( H ) | Classical Complexity | Quantum Complexity |
|---|---|---|---|---|
| Simon's Problem | ( (\mathbb{Z}/2\mathbb{Z})^n ) | ( {0, s} ) | ( \Theta(2^{n/2}) ) [83] | ( O(n) ) [83] |
| Discrete Logarithm | ( \mathbb{Z}{p-1} \times \mathbb{Z}{p-1} ) | ( \langle (s, 1) \rangle ) | Super-polynomial [83] | ( O(\log p) ) [84] |
| Order Finding / Factoring | ( \mathbb{Z} ) | ( r\mathbb{Z} ) (period ( r )) | Super-polynomial [83] | ( O(\log N) ) [84] |
The Researcher's Toolkit: Abelian HSP
Table 4: Essential conceptual tools for the Abelian Hidden Subgroup Problem
| Research Tool | Function in the Algorithm |
|---|---|
| Quantum Oracle for ( f ) | A black-box quantum circuit that implements the function ( f ), which hides the subgroup ( H ) [83]. |
| Quantum Fourier Transform (QFT) | A unitary transformation that reveals the periodic structure (the subgroup ( H )) embedded in a quantum state [84]. |
| Quantum State Tomography | The process of reconstructing the quantum state after the QFT, which is used to identify generators of the subgroup ( H ) [86]. |
| Generalized Fourier Sampling | The core of the "standard method," which samples from the dual group to acquire information about ( H ) [86]. |
Algorithmic Pathway
Comparative Analysis: Quantum vs. Classical Scaling
The experimental data from both domains consistently demonstrates a pattern of quantum advantage rooted in superior scaling laws.
In the chemical simulation of the [4Fe-4S] cluster, the classical computational cost of representing the system's quantum state scales exponentially with electron count. The hybrid approach bypasses this by letting the quantum processor handle the exponentially large search space, while the classical computer solves a refined, smaller problem [27]. While this specific demonstration did not achieve a definitive "quantum advantage" over all classical methods, it points squarely toward that goal. For industrial applications like modeling cytochrome P450 enzymes, estimates suggest that millions of physical qubits may be needed, highlighting the immense scaling challenge that remains [1].
In the Abelian HSP, the contrast is even more stark. Classical algorithms for problems like Simon's require ( \Theta(2^{n/2}) ) queries, while the quantum algorithm requires only ( O(n) ) queries—an exponential speedup [83]. This is not a matter of mere hardware improvement but a fundamental algorithmic divergence. The quantum algorithm's power comes from its ability to exist in a superposition of states, query the function ( f ) once in this superposed state, and then use interference (via the QFT) to extract the global periodicity defined by the subgroup ( H ) [84] [83].
This case study validates the core thesis that quantum computing offers a transformative scaling advantage for specific problem classes critical to chemistry research. The quantum-centric supercomputing study on iron-sulfur clusters provides a tangible, forward-looking blueprint for how hybrid quantum-classical architectures can be deployed today to extract chemically useful information from systems that push the boundaries of classical computation [27]. Simultaneously, the efficient quantum solution to the Abelian HSP provides the mathematical foundation and proof-of-principle for the exponential speedups that are expected to become more prevalent as quantum hardware matures [86] [84] [83].
The path forward is one of co-design: developing quantum algorithms inspired by the HSP framework for specific chemistry problems while advancing hardware to accommodate the demanding requirements of full-scale quantum simulations. The ultimate goal is a self-reinforcing cycle where quantum computers help design better quantum computers, and in doing so, unlock new frontiers in drug discovery, materials science, and our fundamental understanding of molecular interactions.
The process of small-molecule drug discovery is a quintessential example of a large-scale search problem, requiring the navigation of a chemical space estimated to contain over 10^60 potential compounds [87]. Traditional computational methods, while invaluable, operate within a framework of classical computational scaling, where the resources required to simulate molecular systems grow polynomially—and often prohibitively—with system size and complexity. This fundamental limitation is most acute in the accurate simulation of quantum mechanical phenomena, such as non-covalent interactions (NCIs), which are critical for predicting binding affinity but require a level of precision where errors of just 1 kcal/mol can lead to erroneous conclusions [88].
Quantum computing introduces a paradigm shift, offering the potential to overcome these scaling limitations by operating on the very principles that govern molecular behavior. By leveraging quantum bits (qubits) that can exist in superposition, quantum processors can theoretically explore vast molecular configuration spaces simultaneously, rather than sequentially [89] [52]. This review provides a comparative analysis of the emerging performance data for quantum-enhanced drug screening, focusing on the critical metrics of hit rates and operational efficiency that define success in pharmaceutical research. The evidence suggests that hybrid quantum-classical approaches are not merely incremental improvements but are poised to redefine the computational boundaries of chemistry research.
Performance Benchmarking: Quantitative Comparisons
Recent studies and industry reports have begun to quantify the performance advantages of quantum-enhanced screening. The following tables consolidate key comparative data on hit rates, efficiency, and chemical novelty.
Table 1: Comparative Hit Rates and Screening Efficiency
| Screening Approach | Initial Library Size | Compounds Synthesized & Tested | Experimentally Confirmed Hits | Hit Rate | Key Target |
|---|---|---|---|---|---|
| Traditional HTS [90] [91] | 100,000 - 2,000,000 | Thousands to Hundreds of Thousands | Dozens (typical) | ~0.001% - 0.01% | Varies |
| AI-Driven (GALILEO) [90] | 52 Trillion | 12 | 12 | ~100% | Viral RNA Polymerases |
| Quantum-Hybrid (Insilico Medicine) [90] | 100 Million | 15 | 2 | ~13.3% | KRAS-G12D (Oncology) |
Table 2: Computational and Chemical Metrics
| Performance Metric | Traditional / Classical AI | Quantum-Enhanced Approach | Implication |
|---|---|---|---|
| Hit Discovery Rate | Months to Years [91] | Weeks to Months (projected) [90] | Drastically compressed discovery timeline |
| Computational Resource Efficiency | ~40% more parameters required for comparable performance [87] | >60% fewer parameters than classical baseline [87] | More efficient model, reduced computational cost |
| Chemical Novelty (Tanimoto Score) | Lower dissimilarity to known drugs [90] | Higher novelty and diversity [90] [87] | Access to novel, first-in-class chemical matter |
| Binding Affinity | µM to nM range (highly variable) | Sub-µM achieved on difficult targets (e.g., 1.4 µM for KRAS) [90] | Potent activity against previously "undruggable" targets |
Experimental Protocols and Workflows
The superior performance of quantum-enhanced screening stems from fundamentally different computational workflows. Below are the detailed protocols for the key experiments cited in the performance tables.
Protocol: Quantum-Classical Generative Agent (BO-QGAN)
This protocol, derived from the work that achieved a 2.27-fold higher Drug Candidate Score (DCS), outlines a systematic approach to hybrid model architecture [87].
- 1. Molecular Representation: Molecules are represented as graphs ( \mathcal{G(V, E)} ) with up to 9 heavy atoms (C, O, N, F) from the QM9 dataset. Nodes ( vi \in \mathcal{V} ) represent atoms (featured via one-hot encoding), and edges ( (vi, v_j) \in \mathcal{E} ) represent bonds (none, single, double, triple, aromatic). The graph is encoded by a feature matrix ( X \in \mathbb{R}^{N \times T} ) and an adjacency tensor ( A \in \mathbb{R}^{N \times N \times Y} ) [87].
- 2. Hybrid Generator Network (( G_{\theta} )):
- A noise vector ( z ) is embedded into a quantum state using angle encoding.
- A parameterized quantum circuit (PQC) transforms the state. The PQC consists of single-qubit rotations (RY gates) and CNOT-based entangling layers in a ring structure.
- Key architectural finding: The optimal configuration uses 3-4 sequential layers of shallow quantum circuits (4-8 qubits) [87].
- The quantum latent representation is obtained by measuring Pauli-Z expectation values and bridged to a classical neural network.
- 3. Classical Discriminator/Reward Networks: A classical graph neural network (critic) distinguishes real vs. generated molecules. A separate, architecturally identical reward network predicts desirable chemical properties to guide the generator [87].
- 4. Multi-Objective Bayesian Optimization: The quantum circuit's width (M), depth (number of layers), and the classical network's structure are optimized using multi-objective Bayesian optimization to maximize the Drug Candidate Score (DCS) while minimizing parameter count [87].
- 5. Training and Evaluation:
- Loss Function: The generator loss is a weighted sum ( \lambda ) of the adversarial loss from the discriminator and the value loss from the reward network.
- Optimization: Separate Adam optimizers (learning rate ( 1 \times 10^{-4} )) are used for the generator/reward and discriminator networks, with gradient clipping (norm 1.0).
- Validation: Generated molecules are evaluated for realism using Fréchet Distance (FD < 12.5 indicates sufficient realism) and drug-likeness using QED, logP, and Synthetic Accessibility (SA) scores via RDKit [87].
Protocol: Quantum-Enhanced Hit Discovery for KRAS
This protocol details the pipeline used to identify novel inhibitors for the notoriously difficult KRAS-G12D target, achieving a 13.3% experimental hit rate [90].
- 1. Ultra-Large Virtual Screening: The process initiates with the screening of 100 million molecules from a virtual chemical library [90].
- 2. Hybrid Quantum-Classical Filtering: A multi-stage filtering approach is employed:
- Quantum Component: Quantum Circuit Born Machines (QCBMs) are used to explore the vast chemical space and generate molecular structures with high diversity and desired properties [90].
- Classical AI Component: Deep learning models predict molecular properties, binding affinity, and synthesizeability, refining the 100-million-molecule library down to 1.1 million promising candidates [90].
- Performance Note: The hybrid quantum-classical model demonstrated a 21.5% improvement in filtering out non-viable molecules compared to AI-only models [90].
- 3. Synthesis and Experimental Validation: The 1.1 million candidates are further refined to 15 compounds selected for chemical synthesis. These are then tested in biological assays. Two compounds showed confirmed biological activity, one (ISM061-018-2) exhibiting a 1.4 µM binding affinity to KRAS-G12D [90].
Visualizing Workflows and Scaling Relationships
The following diagrams illustrate the core hybrid workflow and the fundamental scaling advantage of quantum approaches.
Hybrid Quantum-Classical Screening Workflow
Computational Scaling in Chemistry
The Scientist's Toolkit: Essential Research Reagents and Solutions
Implementing the protocols above requires a suite of specialized computational tools and platforms. The following table details key resources for building a quantum-enhanced drug discovery pipeline.
Table 3: Key Research Reagent Solutions for Quantum-Enhanced Screening
| Tool / Platform Name | Type | Primary Function in Workflow | Relevance to Performance |
|---|---|---|---|
| GALILEO [90] | Generative AI Platform | Uses deep learning (ChemPrint) for one-shot prediction of novel antiviral compounds. | Achieved 100% hit rate in vitro by expanding chemical space. |
| Quantum Circuit Born Machines (QCBMs) [90] | Quantum Algorithm | Generative models for exploring chemical space and enhancing molecular diversity in hybrid pipelines. | Key for probabilistic modeling, improving molecular diversity in KRAS screen. |
| PennyLane [87] | Software Library | Differentiable programming framework for hybrid quantum-classical machine learning; implements parameterized quantum circuits. | Enables the construction and training of the quantum-classical bridge in optimized GANs. |
| TenCirChem [30] | Quantum Chemistry Package | A software library for efficient simulation of quantum circuits and variational quantum algorithms like VQE. | Facilitates the quantum computation of molecular properties (e.g., energy profiles) in drug design tasks. |
| QUID Benchmark [88] | Dataset/Framework | "QUantum Interacting Dimer" benchmark providing high-accuracy interaction energies for ligand-pocket systems. | Enables calibration and validation of quantum methods against a "platinum standard" for non-covalent interactions. |
| Polarizable Continuum Model (PCM) [30] | Solvation Model | A quantum computational method for modeling solvent effects (e.g., in water) on molecular reactions and properties. | Critical for calculating physiologically relevant Gibbs free energy profiles, as in prodrug activation. |
The comparative data presented in this guide demonstrates that quantum-enhanced drug screening is transitioning from a theoretical promise to a demonstrably powerful tool. The dramatically elevated hit rates—ranging from 13.3% on a high-value oncology target to 100% in an antiviral campaign—coupled with the ability to generate novel chemical matter with high efficiency, signal a profound shift. These performance gains are directly attributable to the superior computational scaling of quantum and hybrid approaches when applied to the intrinsic quantum mechanical problem of molecular simulation. While challenges in quantum hardware stability and scalability remain, the establishment of rigorous benchmarks [88] and reproducible hybrid pipelines [87] [30] provides a clear and objective foundation for researchers to evaluate this transformative technology. The evidence indicates that quantum-enhanced screening is not a distant future prospect but an emerging, high-performance paradigm that is already beginning to redefine the limits of what is computationally possible in chemistry and drug development.
For researchers in chemistry and drug development, the simulation of molecular systems remains a formidable challenge for classical computers. The quantum-mechanical nature of electrons, which dictates molecular structure and reactivity, leads to a computational complexity that scales exponentially with system size, placing fundamental limits on classical computational methods [92]. Quantum computing, which operates on the principles of superposition and entanglement, inherently matches the quantum nature of these problems. It promises to simulate molecular systems with a precision that could revolutionize the discovery of new pharmaceuticals, materials, and catalysts [66] [93]. This guide provides an objective comparison of the current quantum hardware landscape, its projected roadmap, and the experimental data validating its potential for practical impact in chemical research.
The Contending Quantum Hardware Architectures
The race to build a practical quantum computer features several competing hardware approaches, each with distinct strengths and challenges. The following section compares the key players and their architectures.
Comparative Analysis of Major Quantum Hardware
Table 1: Key Hardware Platforms and Specifications
| Company/Entity | Key Processors | Architecture | Key Performance Metrics | Error Correction Milestones |
|---|---|---|---|---|
| Willow (105 qubits) [93] | Superconducting | Completed a benchmark in <5 mins vs. 10²ⵠyears on classical [93]; Demonstrated 13,000x speedup in physics simulation [78] | Achieved exponential error reduction ("below threshold") by scaling qubit arrays [93] | |
| IBM | Heron (133/156 qubits), Nighthawk (120 qubits) [7] [94] | Superconducting with tunable couplers | Nighthawk enables circuits with 30% more complexity, up to 5,000 two-qubit gates [7] | Quantum Loon demonstrates all hardware elements for fault tolerance; real-time decoding 10x faster [7] |
| China (USTC) | Jiuzhang (photonic), Zuchongzhi (66-qubit superconducting) [95] | Photonic & Superconducting | Jiuzhang solved a problem in seconds that would take a supercomputer 600 million years [95] | Actively researching error correction; challenges in qubit connectivity and stability [95] |
| Microsoft | Majorana 1 (in development) [66] | Topological Qubits | Aims for inherent qubit stability with less error correction overhead [66] | Demonstrated 28 logical qubits with a 1,000-fold reduction in error rates [66] |
Roadmaps to Practical Quantum Impact
The hardware development path is structured around clear, ambitious milestones set by leading companies.
- IBM's Path: IBM has outlined one of the most detailed public roadmaps. The company is focusing on achieving quantum advantage—where a quantum computer solves a problem better than all classical methods—by the end of 2026 with its Nighthawk processor and its successors, which are projected to support up to 15,000 two-qubit gates by 2028. The goal is to deliver the first large-scale, fault-tolerant quantum computer by 2029 [7] [94].
- Google's Dual Track: Google is pursuing simultaneous advancements in hardware and software. Following the error correction breakthrough with Willow, the next challenge is to demonstrate a "useful, beyond-classical" computation on a real-world, commercially relevant problem, merging the classical hardness of benchmarks like random circuit sampling with scientific utility [93].
- The Broader Ecosystem: Other players like Atom Computing and Microsoft are also pushing the boundaries of scalability and qubit stability. Microsoft's approach with topological qubits could potentially reduce the monumental overhead typically required for error correction, which is a critical challenge for all other architectures [66].
Experimental Protocols: Benchmarking Quantum Hardware
The claimed capabilities of quantum processors are validated through specific experimental protocols and benchmarks. These experiments provide the critical data for comparing performance across different platforms.
Key Experimental Methodologies
- Random Circuit Sampling (RCS): Pioneered by Google, RCS is a benchmark designed to be exceptionally difficult for classical computers to simulate. The quantum processor executes a series of randomly chosen quantum gates, and its output is sampled. The computational cost for a classical supercomputer to replicate this output distribution is then estimated. Google's Willow processor completed an RCS computation in under five minutes, a task estimated to take the Frontier supercomputer 10 septillion (10²âµ) years [93].
- The Quantum Echoes Algorithm: This protocol, developed by Google Quantum AI, is used to measure a subtle quantum interference phenomenon known as the second-order out-of-time-order correlator (OTOC). The methodology involves four key steps [78]:
- A. Forward Evolution: The quantum system is evolved forward in time.
- B. Butterfly Perturbation: A small, controlled perturbation is applied.
- C. Backward Evolution: The system is evolved backward in time.
- D. Measurement: The interference pattern created by the forward-and-backward evolution is measured, revealing information about quantum chaos and information scrambling. This experiment, run on a 65-qubit processor, was completed in 2.1 hours, a task that would require the Frontier supercomputer approximately 3.2 years, representing a 13,000x speedup [78].
- Quantum Phase Difference Estimation (QPDE): This algorithm, a variant of the foundational Quantum Phase Estimation (QPE) algorithm, is tailored for near-term hardware. In a collaboration between Mitsubishi Chemical Group and Q-CTRL, a tensor-based QPDE algorithm was used to dramatically reduce the resources needed to simulate molecular energy gaps. The experiment successfully ran on an IBM quantum device, reducing the number of CZ gates (a key measure of circuit complexity) from 7,242 to just 794—a 90% reduction—enabling a 5x increase in computational capacity [96].
Experimental Workflow Visualization
The following diagram illustrates the core feedback loop of a hybrid quantum-classical experiment, which is typical for current applications in chemistry research.
Diagram 1: Workflow of a hybrid quantum-classical experiment for chemistry.
The Scientist's Toolkit: Essential Research Reagents & Platforms
Engaging with quantum computing for chemical research requires a suite of software and hardware access platforms. The following table details the key "research reagents" available to scientists today.
Table 2: Essential Tools for Quantum Computational Chemistry
| Tool / Platform | Provider | Function & Utility |
|---|---|---|
| Qiskit | IBM [7] | A full-stack, open-source quantum software development kit (SDK). Its C++ interface and C-API allow integration with high-performance computing (HPC) environments for advanced error mitigation [7]. |
| Fire Opal | Q-CTRL [96] | An AI-powered infrastructure software that automatically handles pulse-level optimization, error suppression, and hardware calibration, enabling higher-fidelity results on today's noisy devices [96]. |
| Quantum Cloud Services (QaaS) | IBM, Google, Microsoft, SpinQ [66] | Cloud-based platforms that provide remote access to quantum processors and simulators, democratizing access and allowing researchers to run experiments without owning hardware [66]. |
| Quantum System Two | IBM [94] | A modular, cryogenic system architecture designed to link multiple quantum processors. It is the cornerstone of IBM's vision for "quantum-centric supercomputing," which integrates quantum and classical resources [94]. |
The hardware roadmap from 100-qubit processors to fault-tolerant machines with millions of qubits is no longer a theoretical exercise but a concerted engineering effort. Current experiments demonstrate that quantum processors are already entering a "beyond-classical" regime for specific tasks, offering speedups that range from thousands to septillions of times for tailored benchmarks [78] [93]. For chemistry research, the recent algorithmic advances, such as QPDE, are rapidly lowering the resource requirements, making meaningful molecular simulations a near-term prospect [96].
The timeline is aggressive, with key milestones like verified quantum advantage targeted for 2026 and fault-tolerant systems by the end of the decade [7]. For researchers and drug development professionals, the time to engage is now. Building expertise in quantum algorithms, leveraging cloud-based quantum resources, and participating in application-focused collaborations are crucial steps to harness the transformative power of quantum computing, which promises to unlock a new era of discovery in chemistry and materials science.
Conclusion
The journey from classical to quantum computational scaling in chemistry is no longer a theoretical pursuit but an emerging reality. The foundational understanding of quantum advantage, combined with methodological advances in hybrid algorithms and successful troubleshooting of noise, has been conclusively validated by recent demonstrations of unconditional exponential speedup. For biomedical and clinical research, this progression signals a paradigm shift. We are moving from the iterative, often inefficient process of guessing and testing molecules toward a future of precise design. The ability to accurately simulate complex biological targets like metalloenzymes and protein-ligand interactions will dramatically accelerate the discovery of novel therapeutics and advanced materials. While challenges in scaling and fault tolerance remain, the trajectory is clear: quantum computing is poised to become an indispensable tool, unlocking a new era of mastery over the molecular world and fundamentally reshaping the landscape of drug discovery and development.
References
- 1. Will quantum computing be chemistry's next AI? - C&EN [https://cen.acs.org/business/quantum-computing-chemistrys-next-AI/103/web/2025/11]
- 2. Kvantify is Scaling Quantum Chemistry on IQM Resonance [https://meetiqm.com/blog/kvantify-is-scaling-quantum-chemistry-on-iqm/]
- 3. Experiments using quantum circuits | Cirq [https://quantumai.google/cirq/experiments]
- 4. New Hybrid Quantum–Classical Computing Approach ... [https://www.caltech.edu/about/news/new-hybrid-quantumclassical-computing-approach-used-to-study-chemical-systems]
- 5. IonQ Quantum Computing Achieves Greater Accuracy ... [https://ionq.com/news/ionq-quantum-computing-achieves-greater-accuracy-simulating-complex-chemical]
- 6. Google Quantum AI Achieves Verifiable ... [https://quantumcomputingreport.com/google-quantum-ai-achieves-verifiable-quantum-advantage-on-willow-chip-with-quantum-echoes-algorithm/]
- 7. IBM Delivers New Quantum Processors, Software, and ... [https://newsroom.ibm.com/2025-11-12-ibm-delivers-new-quantum-processors,-software,-and-algorithm-breakthroughs-on-path-to-advantage-and-fault-tolerance]
- 8. Qiskit - IBM Quantum Computing [https://www.ibm.com/quantum/qiskit]
- 9. Laying the Groundwork for a Quantum Leap in Chemistry [https://www.pnnl.gov/news-media/laying-groundwork-quantum-leap-chemistry]
- 10. Quantitative Description of Strongly Correlated Materials by ... [https://arxiv.org/html/2502.19588v2]
- 11. Density Functional Theory VI: Performance Assessment and ... [https://schedule.aps.org/smt/2025/events/MAR-W67/4]
- 12. A new approach for a quantitative description of strongly ... [https://www.vscentrum.be/post/a-new-approach-for-a-quantitative-description-of-strongly-correlated-materials]
- 13. The strong-interaction limit of density functional theory [https://arxiv.org/abs/2202.09760]
- 14. LK-99 [https://en.wikipedia.org/wiki/LK-99]
- 15. Exploration of superconductivity in LK-99 synthesized ... [https://www.sciencedirect.com/science/article/abs/pii/S1567173924000531]
- 16. 韩国室温超导论文å˜ç¼ºé™·å·²ä¸‹æž¶ï¼å…¨çƒå¤šä¸ªç§‘ç ”å›¢é˜Ÿæµ‹è¯• - 科技 [https://tech.sina.cn/mobile/xp/2023-08-02/detail-imzeutut7446567.d.html?vt=4]
- 17. Absence of Superconductivity in LK-99 at Ambient Conditions [https://pmc.ncbi.nlm.nih.gov/articles/PMC10633996/]
- 18. Leslie Schoop on busting the LK-99 myth - C&EN [https://cen.acs.org/materials/electronic-materials/Leslie-Schoop-debunking-claims-LK-99-room-temperature-superconductor/102/i17]
- 19. It's Back: Researchers Say They've Replicated LK-99 ... [https://thequantuminsider.com/2024/01/04/its-back-researchers-say-theyve-replicated-lk-99-room-temperature-superconductor-experiment/]
- 20. Quantum Computing vs Classical Computing [https://www.spinquanta.com/news-detail/quantum-computing-vs-classical-computing-full-breakdown]
- 21. Quantum Vs. Classical Computing: Understanding ... [https://www.forbes.com/sites/bernardmarr/2025/04/02/quantum-vs-classical-computing-understanding-tomorrows-tech-balance/]
- 22. A Historical Comparison: How Quantum Scaling Differs ... [https://www.linkedin.com/pulse/historical-comparison-how-quantum-scaling-differs-from-classical-n0imf]
- 23. 14 CEOs Break Down the Challenges in Quantum [https://www.louisbouchard.ai/14-ceos-break-down-the-challenges-in-quantum/]
- 24. Quantum vs Classical Computing | Quantum Threat [https://www.quantropi.com/quantum-versus-classical-computing-and-the-quantum-threat/]
- 25. Classical Machine Learning vs Quantum ... [https://chemrxiv.org/engage/chemrxiv/article-details/68e6baea5dd091524f7334f5]
- 26. Quantum Advantage in Computational Chemistry? †[https://arxiv.org/html/2508.20972v1]
- 27. New Hybrid Quantum–Classical Computing Approach Used to ... [https://researchimpact.caltech.edu/research-news/new-hybrid-quantumclassical-computing-approach-used-to-study-chemical-systems]
- 28. Study Suggests Today's Quantum Computers Could Aid ... [https://thequantuminsider.com/2025/07/15/study-suggests-todays-quantum-computers-could-aid-molecular-simulation/]
- 29. Hybrid Quantum–Classical Machine Learning Potential ... [https://arxiv.org/html/2508.04098v1]
- 30. A hybrid quantum computing pipeline for real world drug ... [https://www.nature.com/articles/s41598-024-67897-8]
- 31. Evaluating the performance of classical, quantum, and ... [https://www.nature.com/articles/s41598-025-22950-y]
- 32. pnnl/QASMBench: A low-level OpenQASM benchmark ... [https://github.com/pnnl/QASMBench]
- 33. BenchQC: A Benchmarking Toolkit for Quantum Computation [https://arxiv.org/html/2502.09595v1]
- 34. A study on Quantum Computing Could Revolutionize Drug ... [https://www.ijsrtjournal.com/article/A-study-on-Quantum-Computing-Could-Revolutionize-Drug-Discovery]
- 35. Variational Quantum Eigensolver: A Comparative Analysis ... [https://arxiv.org/html/2412.19176v2]
- 36. Greedy Quantum Algorithm Charts a New Course to ... [https://blog.qubit-pharmaceuticals.com/blog/greedy-quantum-algorithm-charts-a-new-course-to-ground-state-energy-success-1]
- 37. BenchQC: A Benchmarking Toolkit for Quantum Computation [https://pmc.ncbi.nlm.nih.gov/articles/PMC12614196/]
- 38. BenchQC: A Benchmarking Toolkit for Quantum Computation [https://www.nist.gov/publications/benchqc-benchmarking-toolkit-quantum-computation]
- 39. Benchmarking VQE Configurations Enables Silicon ... [https://quantumzeitgeist.com/vqe-variational-quantum-benchmarking-configurations-enables-silicon-ground-state-energy-estimation/]
- 40. Comparative study of quantum and classical algorithms for ... [https://www.sciencedirect.com/science/article/pii/S2590123025031172]
- 41. New World-Record in Quantum Volume [https://www.quantinuum.com/blog/quantum-volume-milestone]
- 42. KRAS: Biology, Inhibition, and Mechanisms of Inhibitor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11049385/]
- 43. Advances in biomarkers of resistance to KRAS mutation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12508421/]
- 44. Quantum-computing-enhanced algorithm unveils potential ... [https://www.nature.com/articles/s41587-024-02526-3]
- 45. Quantum Computing in Drug Discovery Techniques ... [https://pubmed.ncbi.nlm.nih.gov/40873222/]
- 46. QM/MM study reveals novel mechanism of KRAS and ... [https://www.sciencedirect.com/science/article/abs/pii/S0141813025003691]
- 47. Identification of New KRAS G12D Inhibitors through ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8836163/]
- 48. quantum computing accelerates KRAS inhibitor design - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12075789/]
- 49. QKDTI A quantum kernel based machine learning model ... [https://www.nature.com/articles/s41598-025-07303-z]
- 50. Scaling Quantum Computing Research to a New Milestone [https://www.ansys.com/blog/scaling-quantum-computing-research]
- 51. [2509.14470] Scaling Hybrid Quantum-HPC Applications ... [https://arxiv.org/abs/2509.14470]
- 52. How quantum is changing molecular drug development computing [https://www.weforum.org/stories/2025/01/quantum-computing-drug-development/]
- 53. Streamlining Protein Folding for Quantum Computers [https://qoroquantum.net/?news=streamlining-protein-folding-quantum-computers-protein-folding]
- 54. Researchers Use Trapped-Ion Quantum Computer to ... [https://thequantuminsider.com/2025/06/15/researchers-use-trapped-ion-quantum-computer-to-tackle-tricky-protein-folding-problems/]
- 55. IonQ and Kipu Quantum Break New Performance Records For... [https://thequantuminsider.com/2025/06/20/ionq-and-kipu-quantum-break-new-performance-records-for-protein-folding-and-optimization-problems/]
- 56. with an all-to-all trapped-ion Protein folding quantum computer [https://arxiv.org/abs/2506.07866]
- 57. Quantum computing and chemistry [https://www.sciencedirect.com/science/article/pii/S2666386424003837]
- 58. Dynamical decoupling for superconducting qubits [https://arxiv.org/html/2207.03670v3]
- 59. Mitigating Idling Errors in Qubits via Adaptive Dynamical ... [https://arxiv.org/abs/2109.05309]
- 60. Measuring error rates of mid-circuit measurements [https://www.nature.com/articles/s41467-025-60923-x]
- 61. Multireference error mitigation for quantum computation ... [https://pubs.rsc.org/en/content/articlelanding/2025/dd/d5dd00202h]
- 62. Quantum Report Says Error Correction Now The Industry's ... [https://thequantuminsider.com/2025/11/19/quantum-report-says-error-correction-now-the-industrys-defining-challenge/]
- 63. A potential quantum leap [https://news.harvard.edu/gazette/story/2025/11/a-potential-quantum-leap/]
- 64. Alice & Bob Accelerates Fault-Tolerant Quantum ... [https://alice-bob.com/newsroom/alice-bob-accelerates-fault-tolerant-quantum-computing-with-nvidia-nvqlink/]
- 65. QEC25 [https://qec25.yalepages.org/]
- 66. Quantum Computing Industry Trends 2025: A Year of ... [https://www.spinquanta.com/news-detail/quantum-computing-industry-trends-2025-breakthrough-milestones-commercial-transition]
- 67. Researchers Find Classical Methods Will Likely ... [https://quantumzeitgeist.com/researchers-find-classical-methods-outperform-algorithms-for-at-least-the-next-two-decades-in-computational-chemistry/]
- 68. A general framework for active space embedding methods ... [https://www.nature.com/articles/s41524-024-01477-2]
- 69. Circuit Optimization And The Quantum Transpilation Problem [https://quantumzeitgeist.com/circuit-optimization-and-the-quantum-transpilation-problem/]
- 70. Quantum computational chemistry [https://en.wikipedia.org/wiki/Quantum_computational_chemistry]
- 71. Resource estimation for quantum chemistry [https://pennylane.ai/qml/demos/tutorial_resource_estimation]
- 72. Quantum Information-Assisted Complete Active Space ... [https://arxiv.org/html/2309.01676v2]
- 73. Quantum vs. classical: A comprehensive benchmark study ... [https://arxiv.org/abs/2504.12416]
- 74. Benchmarking near-term quantum algorithms is an art [https://pennylane.ai/blog/2024/03/benchmarking-near-term-quantum-algorithms-is-an-art]
- 75. Quantum-classical hybrid algorithm for solving the learning ... [https://www.nature.com/articles/s42005-025-02126-w]
- 76. New USC study demonstrates unconditional exponential ... [https://viterbischool.usc.edu/news/2025/06/new-usc-study-demonstrates-unconditional-exponential-quantum-scaling-advantage/]
- 77. A Perspective on Quantum Computing Applications in ... [https://arxiv.org/html/2506.19337v2]
- 78. Google Quantum AI Shows 13000× Speedup Over World's ... [https://thequantuminsider.com/2025/10/22/google-quantum-ai-shows-13000x-speedup-over-worlds-fastest-supercomputer-in-physics-simulation/]
- 79. Top quantum breakthroughs of 2025 [https://www.networkworld.com/article/4088709/top-quantum-breakthroughs-of-2025.html]
- 80. One small qubit, one giant leap for quantum computing [https://www.sciencedaily.com/releases/2025/07/250724040459.htm]
- 81. Experts See a Technological Turf Battle Brewing Between ... [https://thequantuminsider.com/2024/11/08/experts-see-a-technological-turf-battle-brewing-between-quantum-computing-and-classical-ai-in-chemistry-and-materials-science/]
- 82. Iron-sulfur clusters: the road to room temperature - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11928408/]
- 83. The Hidden Subgroup Problem [https://www.daniellowengrub.com/blog/2025/04/23/hidden-subgroup]
- 84. Hidden subgroup problem [https://en.wikipedia.org/wiki/Hidden_subgroup_problem]
- 85. [2507.18088] An Initialization-free Quantum Algorithm for ... [https://arxiv.org/abs/2507.18088]
- 86. [2505.15770] The abelian state hidden subgroup problem [https://arxiv.org/abs/2505.15770]
- 87. Bridging Quantum and Classical Computing in Drug Design [https://arxiv.org/html/2506.01177v1]
- 88. Extending quantum-mechanical benchmark accuracy to ... [https://www.nature.com/articles/s41467-025-63587-9]
- 89. The Billion-Dollar Bet That Could Rewrite Drug Discovery [https://thedigitalleader.substack.com/p/quantum-gambit-the-billion-dollar]
- 90. The Future of Drug Discovery: 2025 as the Inflection Year ... [https://modelmedicines.com/newsroom/the-future-of-drug-discovery-2025-as-the-inflection-year-for-hybrid-ai-and-quantum-computing]
- 91. Computational approaches streamlining drug discovery [https://www.nature.com/articles/s41586-023-05905-z]
- 92. Google, IBM make strides toward quantum computers that ... [https://www.cbsnews.com/news/quantum-computing-google-ibm-advances-60-minutes-transcript/]
- 93. Meet Willow, our state-of-the-art quantum chip [https://blog.google/technology/research/google-willow-quantum-chip/]
- 94. IBM Quantum Computing | Hardware and roadmap [https://www.ibm.com/quantum/hardware]
- 95. Google's, IBM's, and China's Quantum Computers [https://www.linkedin.com/pulse/googles-ibms-chinas-quantum-computers-comparative-analysis-ansal-mt-l2erc]
- 96. Unlocking new performance capability in quantum chemistry [https://q-ctrl.com/case-study/unlocking-new-performance-capability-in-quantum-chemistry]